













| General Information: |
|
| Name(s) found: |
EF109_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 268 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
|
| Biological Process: |
regulation of transcription, DNA-dependent
[ISS]
response to chitin [IEP] defense response to fungus [IEP] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHYPNNRTEF VGAPAPTRYQ KEQLSPEQEL SVIVSALQHV ISGENETAPC QGFSSDSTVI 60
61 SAGMPRLDSD TCQVCRIEGC LGCNYFFAPN QRIEKNHQQE EEITSSSNRR RESSPVAKKA 120
121 EGGGKIRKRK NKKNGYRGVR QRPWGKFAAE IRDPKRATRV WLGTFETAED AARAYDRAAI 180
181 GFRGPRAKLN FPFVDYTSSV SSPVAADDIG AKASASASVS ATDSVEAEQW NGGGGDCNME 240
241 EWMNMMMMMD FGNGDSSDSG NTIADMFQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
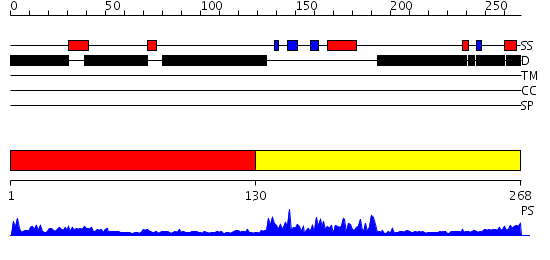
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..129] | 1.130995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [130..268] | 22.0 | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)