













| General Information: |
|
| Name(s) found: |
APS4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 469 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrion [IDA] |
| Biological Process: |
sulfate assimilation
[TAS]
|
| Molecular Function: |
sulfate adenylyltransferase (ATP) activity
[IMP][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASSAAAIVS GSPFRSSPLI HNHHASRYAP GSISVVSLPR QVSRRGLSVK SGLIEPDGGK 60
61 LMNLVVEESR RRVMKHEAET VPARIKLNRV DLEWVHVLSE GWASPLKGFM RQSEFLQTLH 120
121 FNSFRLEDGS VVNMSVPIVL AIDDDQKFRI GDSNQVTLVD SVGNPIAILN DIEIYKHPKE 180
181 ERIARTWGTT ARGLPYAEEA ITKAGNWLIG GDLQVLEPIK YNDGLDRFRL SPSQLREEFI 240
241 RRGADAVFAF QLRNPVHNGH ALLMTDTRRR LLEMGYKNPV LLLNPLGGFT KADDVPLSWR 300
301 MRQHEKVLED GVLDPETTVV SIFPSPMLYA GPTEVQWHAK ARINAGANFY IVGRDPAGMG 360
361 HPTEKRDLYD ADHGKKVLSM APGLERLNIL PFKVAAYDKT QGKMAFFDPS RSQDFLFISG 420
421 TKMRGLAKKK ENPPDGFMCP SGWKVLVDYY DSLSAETGNG RVSEAVASA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
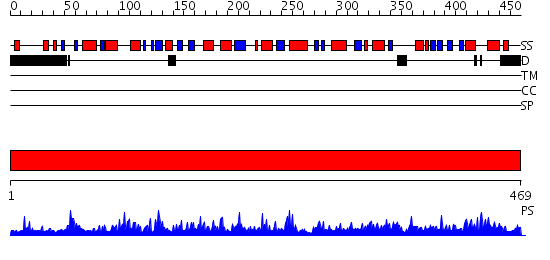
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..469] | 156.0 | The crystal structure of human 3'-phosphoadenosine-5'-phosphosulfate synthetase 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.58 |
Source: Reynolds et al. (2008)