













| General Information: |
|
| Name(s) found: |
AP3D_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 869 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane coat
[IEA]
Golgi apparatus [IEA] |
| Biological Process: |
vesicle-mediated transport
[IEA]
protein transport [IEA] intracellular protein transport [IEA] |
| Molecular Function: |
protein binding
[IEA]
protein transporter activity [IEA] clathrin binding [ISS] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSSTSIMD NLFQRSLEDL IKGFRLQLLG ESNFISRAVE EIRREIKATD LSTKSTALHK 60
61 LSYLAALHGV DMSWAAFHAV EVVSSSRFQH KRIGYQAITQ SFNDQTSVML LITNQVRKDL 120
121 NSANEYEVSL ALECLSRIGT HDLARDLTPE VFTLLGSSKS FVKKKAIGVV LRVFEKYHDA 180
181 VKVCFKRLVE NLETSDPQIL SAVVGVFCEL ATKDPQSCLP LAPEFYKVLV DSRNNWVLIK 240
241 VLKIFAKLAL IEPRLGKKVA EPICEHMRRT VAKSLVFECV RTVVSSLSDN EAAVKLAVAK 300
301 IREFLVEDDP NLKYLGLNAL SIVAPKHLWA VLENKEVVVK AMSDEDPNVK LEALHLLMAM 360
361 VNEDNVSEIS RILMNYALKS DPLFCNEIIF SVLSACSRNA YEIIVDFDWY LSLLGEMARI 420
421 PHCQRGEDIE HQLIDIGMRV RDARPQLVRV SWALLIDPAL LGNLFLHPIL SAAAWVSGEY 480
481 VEFSKNPYET VEALLQPRTD LLPPSIKAIY IHSAFKVLVF CLGSYFSSQE PTSSSLAQES 540
541 SSGSLLVNVF THESILSLVN VIELGLGPLS GYHDVEVQER AKNVLGYISV IKQEIAEQLN 600
601 LQDNETEASR VTAFMEDVFS EEFGPISATA QEKVCVPDGL ELKENLGDLE EICGEHLKPV 660
661 ESDSVSYTDK ISFSVSKLRI RDQQEATSSS SPPHEASSLL AEHRKRHGMY YLTSQKEDQD 720
721 SNGTSSDYPL ANELANEISQ DSFNPKRKPN QSKPRPVVVK LDDGDESRIT PQAKTNIQTA 780
781 NDDESLSRAI QSALLVKNKG KEKDRYEGNP NSGQQEKEES SRIENHQNSE KKKKKKKKKK 840
841 GEGSSKHKSR RQNEVASASE QVIIPDFLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
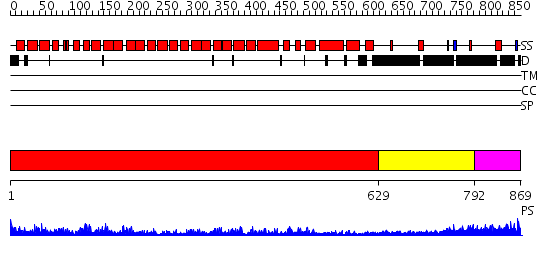
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..628] | 91.69897 | AP1 clathrin adaptor core | |
| 2 | View Details | [629..791] | 7.522879 | Gamma1-adaptin domain | |
| 3 | View Details | [792..869] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.75 |
Source: Reynolds et al. (2008)