













| General Information: |
|
| Name(s) found: |
CMT1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 791 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
chromatin [IEA] |
| Biological Process: |
DNA mediated transformation
[IMP]
DNA methylation [ISS] |
| Molecular Function: |
DNA binding
[IEA][ISS]
chromatin binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAARNKQKKR AEPESDLCFA GKPMSVVEST IRWPHRYQSK KTKLQAPTKK PANKGGKKED 60
61 EEIIKQAKCH FDKALVDGVL INLNDDVYVT GLPGKLKFIA KVIELFEADD GVPYCRFRWY 120
121 YRPEDTLIER FSHLVQPKRV FLSNDENDNP LTCIWSKVNI AKVPLPKITS RIEQRVIPPC 180
181 DYYYDMKYEV PYLNFTSADD GSDASSSLSS DSALNCFENL HKDEKFLLDL YSGCGAMSTG 240
241 FCMGASISGV KLITKWSVDI NKFACDSLKL NHPETEVRNE AAEDFLALLK EWKRLCEKFS 300
301 LVSSTEPVES ISELEDEEVE ENDDIDEAST GAELEPGEFE VEKFLGIMFG DPQGTGEKTL 360
361 QLMVRWKGYN SSYDTWEPYS GLGNCKEKLK EYVIDGFKSH LLPLPGTVYT VCGGPPCQGI 420
421 SGYNRYRNNE APLEDQKNQQ LLVFLDIIDF LKPNYVLMEN VVDLLRFSKG FLARHAVASF 480
481 VAMNYQTRLG MMAAGSYGLP QLRNRVFLWA AQPSEKLPPY PLPTHEVAKK FNTPKEFKDL 540
541 QVGRIQMEFL KLDNALTLAD AISDLPPVTN YVANDVMDYN DAAPKTEFEN FISLKRSETL 600
601 LPAFGGDPTR RLFDHQPLVL GDDDLERVSY IPKQKGANYR DMPGVLVHNN KAEINPRFRA 660
661 KLKSGKNVVP AYAISFIKGK SKKPFGRLWG DEIVNTVVTR AEPHNQCVIH PMQNRVLSVR 720
721 ENARLQGFPD CYKLCGTIKE KYIQVGNAVA VPVGVALGYA FGMASQGLTD DEPVIKLPFK 780
781 YPECMQAKDQ I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
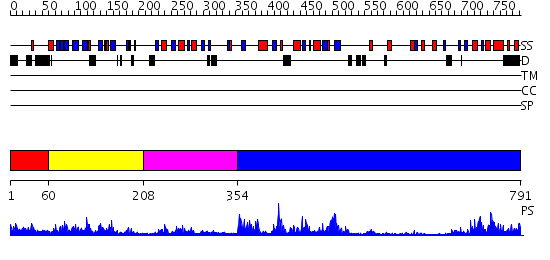
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..59] | 1.105996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [60..207] | 28.39794 | Crystal structure of the proximal BAH domain of polybromo | |
| 3 | View Details | [208..353] | 4.99 | DNA methylase HaeIII | |
| 4 | View Details | [354..791] | 42.0 | DNMT2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)