













| General Information: |
|
| Name(s) found: |
ZEP_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 667 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
chloroplast envelope [IDA] |
| Biological Process: |
response to heat
[IMP]
sugar mediated signaling pathway [TAS] response to osmotic stress [IMP] abscisic acid biosynthetic process [TAS] xanthophyll biosynthetic process [IMP] response to red light [IEP] response to water deprivation [IMP] |
| Molecular Function: |
zeaxanthin epoxidase activity
[IMP][TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSTPFCYSI NPSPSKLDFT RTHVFSPVSK QFYLDLSSFS GKPGGVSGFR SRRALLGVKA 60
61 ATALVEKEEK REAVTEKKKK SRVLVAGGGI GGLVFALAAK KKGFDVLVFE KDLSAIRGEG 120
121 KYRGPIQIQS NALAALEAID IEVAEQVMEA GCITGDRING LVDGISGTWY VKFDTFTPAA 180
181 SRGLPVTRVI SRMTLQQILA RAVGEDVIRN ESNVVDFEDS GDKVTVVLEN GQRYEGDLLV 240
241 GADGIWSKVR NNLFGRSEAT YSGYTCYTGI ADFIPADIES VGYRVFLGHK QYFVSSDVGG 300
301 GKMQWYAFHE EPAGGADAPN GMKKRLFEIF DGWCDNVLDL LHATEEEAIL RRDIYDRSPG 360
361 FTWGKGRVTL LGDSIHAMQP NMGQGGCMAI EDSFQLALEL DEAWKQSVET TTPVDVVSSL 420
421 KRYEESRRLR VAIIHAMARM AAIMASTYKA YLGVGLGPLS FLTKFRVPHP GRVGGRFFVD 480
481 IAMPSMLDWV LGGNSEKLQG RPPSCRLTDK ADDRLREWFE DDDALERTIK GEWYLIPHGD 540
541 DCCVSETLCL TKDEDQPCIV GSEPDQDFPG MRIVIPSSQV SKMHARVIYK DGAFFLMDLR 600
601 SEHGTYVTDN EGRRYRATPN FPARFRSSDI IEFGSDKKAA FRVKVIRKTP KSTRKNESNN 660
661 DKLLQTA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
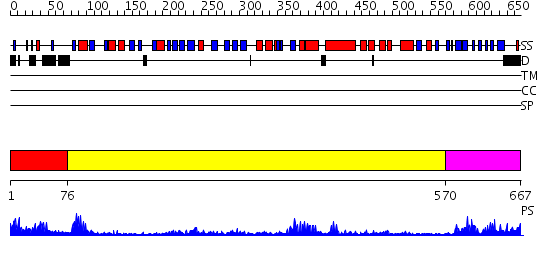
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | 13.39794 | Crystal structure of the native monooxygenase domain of MICAL at 1.45 A resolution | |
| 2 | View Details | [76..569] | 41.221849 | No description for 2dkhA was found. | |
| 3 | View Details | [570..667] | 9.154902 | The solution structure of the Ki67FHA/hNIFK(226-269)3P complex |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.56 |
Source: Reynolds et al. (2008)