













| General Information: |
|
| Name(s) found: |
PI5K2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 754 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
reproductive developmental process
[IMP]
growth [IMP] |
| Molecular Function: |
1-phosphatidylinositol-4-phosphate 5-kinase activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMREPLVSEE EEEEATEVLL VEKTKLCKRR GDEEKTEERR DDLLLLALTP MVRSKSQGTT 60
61 RRVTPTPPPV DVEKPLPNGD LYMGTFSGGF PNGSGKYLWK DGCMYEGEWK RGKASGKGKF 120
121 SWPSGATYEG EFKSGRMEGS GTFVGVDGDT YRGSWVADRK QGHGQKRYAN GDYYEGTWRR 180
181 NLQDGRGRYV WMNGNQYTGE WRNGVICGKG VLAWPNGNRY EGQWENGVPK GSGVFTWADG 240
241 SSWIGSWNES SNLMRNFFDG IEKNELIVAT RKRSSVDSGA GSLTGEKIFP RICIWESDGE 300
301 AGDITCDIVD NVEASVIYRD RISIDKDGFR QFRKNPCCFS GEAKKPGETI SKGHKKYDLM 360
361 LNLQHGIRYS VGKHASVVRD LKQSDFDPSE KFWTRFPPEG SKTTPPHLSV DFRWKDYCPL 420
421 VFRRLRELFT VDPADYMLAI CGNDALRELS SPGKSGSFFY LTQDDRFMIK TVKKSEVKVL 480
481 LRMLPSYYKH VCQYENTLVT RFYGVHCIKP VGGQKTRFIV MGNLFCSEYR IQRRFDLKGS 540
541 SHGRYTSKPE GEIDETTTLK DLDLNFAFRL QRNWYQELMT QIKRDCEFLE AERIMDYSLL 600
601 VGVHFRDDNT GDKMGLSPFV LRSGKIESYQ SEKFMRGCRF LEAELQDMDR ILAGRKPLIR 660
661 LGANMPARAE RMARRSDYDQ YSSGGTNYQS HGEVYEVVLY FGIIDILQDY DISKKIEHAY 720
721 KSLQADPASI SAVDPKLYSR RFRDFISRIF IEDG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
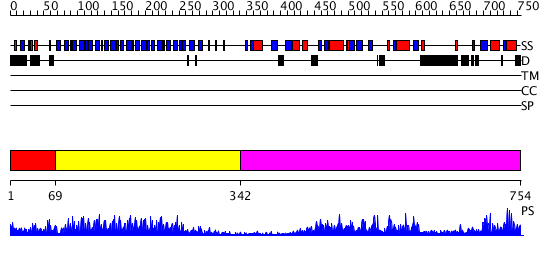
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..68] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [69..341] | 7.522879 | Crystal structure of the complete modular teichioic acid phosphorylcholine esterase Pce (CbpE) from Streptococcus pneumoniae | |
| 3 | View Details | [342..754] | 88.39794 | Phosphatidylinositol phosphate kinase IIbeta, PIPK IIbeta |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)