













| General Information: |
|
| Name(s) found: |
gi|5734747
[NCBI NR]
gi|25513500 [NCBI NR] gi|15220054 [NCBI NR] gi|110738070 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 958 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytosol [IDA] phragmoplast [IDA] |
| Biological Process: | NONE FOUND |
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKEEDVSSQN VNPRSNRNSV ASASASASAT PVDRFRRRAR SPSPPQTAAA SSAGASSPAV 60
61 LVNAGSVDWT GHGLALSVRS CRTWDRGDLL RRLATFKPSN WLGKPKTASS LACAQKGWVS 120
121 VDLDKLQCEY CGSILQYSPP QDSLNPPEAD TTGEKFSKQL DDAHESSCPW VGKSCSESLV 180
181 QFPPTPPSAL IGGYKDRCDG LLQFYSLPIV SPSAIDQMRA SRRPQIDRLL AHANDDLSFR 240
241 MDNISAAETY KEEAFSNYSR AQKLISLCGW EPRWLPNIQD CEEHSAQSAR NGCPSGPARN 300
301 QSRLQDPGPS RKQFSASSRK ASGNYEVLGP EYKSESRLPL LDCSLCGVTV RICDFMTTSR 360
361 PVPFAAINAN LPETSKKMGV TRGTSATSGI NGWFANEGMG QQQNEDVDEA ETSVKRRLVS 420
421 NVGLSFYQNA AGASSSAQLN MSVTRDNYQF SDRGKEVLWR QPSGSEVGDR AASYESRGPS 480
481 TRKRSLDDGG STVDRPYLRI QRADSVEGTV VDRDGDEVND DSAGPSKRTR GSDAHEAYPF 540
541 LYGRDLSVGG PSHSLDAENE REVNRSDPFS EGNEQVMAFP GARDSTRASS VIAMDTICHS 600
601 ANDDSMESVE NHPGDFDDIN YPSVATAQSA DFNDPSELNF SNQAQQSACF QPAPVRFNAE 660
661 QGISSINDGE EVLNTETVTA QGRDGPSLGV SGGSVGMGAS HEAEIHGADV SVHRGDSVVG 720
721 DMEPVAEVIE NLGQSGEFAP DQGLTDDFVP AEMDREGRLG DSQDRVSQSV VRADSGSKIV 780
781 DSLKAESVES GEKMSNINVL INDDSVHPSL SCNAIVCSGY EASKEEVTQT WESPLNAGFA 840
841 LPGSSYTAND QGPQNGDSND DIVEFDPIKY HNCYCPWVNE NVAAAGCSSN SSGSSGFAEA 900
901 VCGWQLTLDA LDSFQSLENP QNQTMESESA ASLCKDDHRT PSQKLLKRHS FISSHGKK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
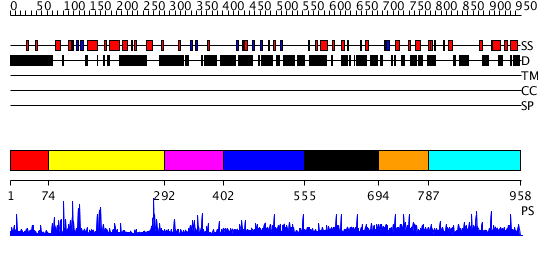
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..73] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [74..291] | 62.154902 | No description for PF07967.5 was found. No confident structure predictions are available. | |
| 3 | View Details | [292..401] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [402..554] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [555..693] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [694..786] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [787..958] | 1.004936 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)