













| General Information: |
|
| Name(s) found: |
PKP4_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 710 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
glycolysis
[IEA][ISS]
|
| Molecular Function: |
magnesium ion binding
[IEA]
pyruvate kinase activity [IEA][ISS] potassium ion binding [IEA] catalytic activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVGLDSSHLL RDKILCFSSR SHINNQHKKT SYALSLNHMK LPIQRTLAFA LARGKGEAES 60
61 FSRLEATFGD NTSTECTWSF DFPDSKDAMS HLKSEADLSG SNGANNVASV IEKLNALRSH 120
121 LLAAEKWNAS QLHLCDSKYL ECATNLVHYM ALRSLDIEQL NSHLASLGLS SLDNNNLDVL 180
181 AHLNASINLL MNDQNAVTES WTNVYPKGKS TKKNDKGRVL SYKESLLGKL REGRSTHIMV 240
241 TIGEEATLSE TFITDILKAG TSVIRINCAH GDPSIWGEII KRVRRTSQML EMPCRVHMDL 300
301 AGPKLRTGTL KPGPCVMKIS PKKDAYGNVV SPALVWLCLT GTEPPAHVSP DATISVQGQD 360
361 FLAGLQIGDS IRLCDARGRK RRLKISKEFH VFNSTGFVAE CFDTAYIESG TELSVKGKKG 420
421 RRLVGRVVDV PPKESFVRLK VGDLLVITRE GSLDEPSVTV PGAHRLTCPS GYLFDSVKPG 480
481 ETIGFDDGKI WGVIKGTSPS EVIVSITHAR PKGTKLGSEK SINIPQSDIH FKGLTSKDIK 540
541 DLDYVASHAD MVGISFIRDV HDITVLRQEL KKRKLDDLGI VLKIETKSGF KNLSLILLEA 600
601 MKCSNPLGIM IARGDLAVEC GWERLANMQE EIIAICKAAR VPVIMATQVL ESLVKSGVPT 660
661 RAEITDAANA KRASCVMLNK GKNIVEAVSM LDTILHTKLI YKKSDSENLH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
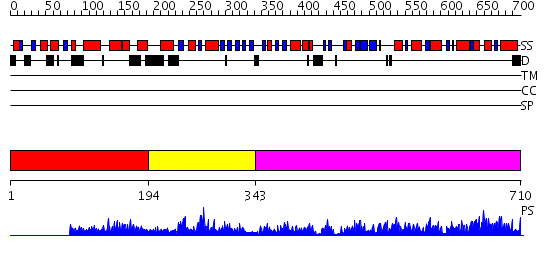
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..193] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [194..342] | 2.6 | Pyruvate kinase (PK); Pyruvate kinase, N-terminal domain; Pyruvate kinase, C-terminal domain | |
| 3 | View Details | [343..710] | 126.0 | Structure of human muscle pyruvate kinase (PKM2) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)