













| General Information: |
|
| Name(s) found: |
DNJA6_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 442 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
chloroplast thylakoid membrane [IDA] |
| Biological Process: |
response to heat
[IEA]
protein folding [IEA][ISS] |
| Molecular Function: |
heat shock protein binding
[IEA]
ATP binding [IEA] unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAIIQLGSTC VAQWSIRPQF AVRAYYPSRI ESTRHQNSSS QVNCLGASKS SMFSHGSLPF 60
61 LSMTGMSRNM HPPRRGSRFT VRADADYYSV LGVSKNATKA EIKSAYRKLA RNYHPDVNKD 120
121 PGAEEKFKEI SNAYEVLSDD EKKSLYDRYG EAGLKGAAGF GNGDFSNPFD LFDSLFEGFG 180
181 GGMGRGSRSR AVDGQDEYYT LILNFKEAVF GMEKEIEISR LESCGTCEGS GAKPGTKPTK 240
241 CTTCGGQGQV VSAARTPLGV FQQVMTCSSC NGTGEISTPC GTCSGDGRVR KTKRISLKVP 300
301 AGVDSGSRLR VRGEGNAGKR GGSPGDLFVV IEVIPDPILK RDDTNILYTC KISYIDAILG 360
361 TTLKVPTVDG TVDLKVPAGT QPSTTLVMAK KGVPVLNKSN MRGDQLVRVQ VEIPKRLSKE 420
421 EKKLIEELAD MSKNKTANST SR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
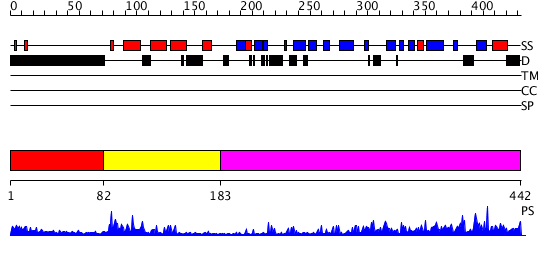
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..81] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [82..182] | 19.69897 | J-DOMAIN (RESIDUES 1-77) OF THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 1-104) OF THE MOLECULAR CHAPERONE DNAJ, NMR, 20 STRUCTURES | |
| 3 | View Details | [183..442] | 46.0 | The crystal structure of Hsp40 Ydj1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)