













| General Information: |
|
| Name(s) found: |
LOX6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 917 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
growth
[ISS]
|
| Molecular Function: |
oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen
[IEA]
iron ion binding [IEA] metal ion binding [IEA] electron carrier activity [IEA] lipoxygenase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFVASPVKTN FNGVSLVKSP AFSALSCRKQ HRVPISRQVR AVISREEKAV DQEDGKKSTN 60
61 KPLINSSQFP WQRSKYTGSK TVTAVVKIRK KIKEKLTERF EHQLELFMKA IGQGMLIQLV 120
121 SEEIDPETGK GRKSLESPVM GLPKAVKDPR YLVFTADFTV PINFGKPGAI LVTNLLSTEI 180
181 CLSEIIIEDS TDTILFPANT WIHSKNDNPQ ARIIFRSQPC LPSETPDGIK ELREKDLVSV 240
241 RGDGKGERKP HERIYDYDVY NDLGDPRKTE RVRPVLGVPE TPYPRRCRTG RPLVSKDPPC 300
301 ESRGKEKEEF YVPRDEVFEE IKRDTFRAGR FKALFHNLVP SIAAALSNLD IPFTCFSDID 360
361 NLYKSNIVLG HTEPKDTGLG GFIGGFMNGI LNVTETLLKY DTPAVIKWDR FAWLRDNEFG 420
421 RQALAGVNPV NIELLKELPI RSNLDPALYG PQESVLTEEI IAREVEHYGT TIEKALEEKR 480
481 LFLVDYHDIL LPFVEKINSI KEDPRKTYAS RTIFFYSKNG ALRPLAIELS LPPTAESENK 540
541 FVYTHGHDAT THWIWKLAKA HVCSNDAGVH QLVNHWLRTH ASMEPYIIAT NRQLSTMHPV 600
601 YKLLHPHMRY TLEINARARK SLINGGGIIE SCFTPGKYAM ELSSAAYKSM WRFDMEGLPA 660
661 DLVRRGMAEE DSSAECGVRL VIDDYPYAAD GLLIWKAIKD LVESYVKHFY SDSKSITSDL 720
721 ELQAWWDEIK NKGHYDKKDE PWWPKLNTTQ DLSQILTNMI WIASGQHAAI NFGQYPFGGY 780
781 VPNRPTLLRK LIPQETDPDY EMFMRNPQYS FLGSLPTQLQ ATKVMAVQET LSTHSPDEEY 840
841 LIELREVQRH WFQDEQVVKY FNKFSEELVK IEKTINERNK DKKLKNRTGA GMPPYELLLP 900
901 TSPHGVTGRG IPNSISI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
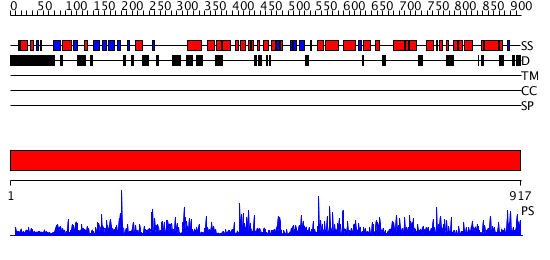
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..917] | 1000.0 | Lipoxigenase, C-terminal domain; Plant lipoxigenase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)