













| General Information: |
|
| Name(s) found: |
gi|9758212
[NCBI NR]
gi|30696661 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 682 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[ISS]
|
| Biological Process: |
cell proliferation
[IMP]
leaf morphogenesis [IMP] root morphogenesis [IMP] |
| Molecular Function: |
RNA binding
[IEA]
S-adenosylmethionine-dependent methyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPALTRNKKK AATKSITPPT KQLTKSKTPP MKPQTSMLKK GAKSQNKPPL KKQKKEVVEE 60
61 EPLEDYEVTD DSDEDDEVSD GSDEDDISPA VESEEIDESD DGENGSNQLF SDDEEENDEE 120
121 TLGDDFLEGS GDEDEEGSLD ADSDADSDDD DIVAKSDAID RDLAMQKKDA AAELEDFIKQ 180
181 DDVHDEEPEH DAFRLPTEEE LEEEARGPPD LPLLKTRIEE IVRALKNFKA FRPKDTTRKA 240
241 CVEQLKADLG SYYGYNSFLI GTLVEMFPPG ELMELIEAFE KQRPTSIRTN TLKTRRRDLA 300
301 DVLLNRGVNL DPLSKWSKVG LVIYDSQVPI GATPEYLAGY YMLQGASSFL PVMALAPREN 360
361 ERIVDVAAAP GGKTTYIAAL MKNTGLIYAN EMKVPRLKSL TANLHRMGVT NTIVCNYDGR 420
421 ELPKVLGQNT VDRVLLDAPC SGTGIISKDE SVKITKTMDE IKKFAHLQKQ LLLAAIDMVD 480
481 ANSKTGGYIV YSTCSIMVTE NEAVIDYALK KRDVKLVTCG LDFGRKGFTR FREHRFQPSL 540
541 DKTRRFYPHV HNMDGFFVAK LKKMSNVKQS SEEGDDDAVE TVEQAEVSSD DDDEAEAIEE 600
601 TEKPSVPVRQ PKERKEKKNK EKLAKSKEDK RGKKDKKSKS ENVEEPSKPR KQKKKRREWK 660
661 NEIAQAREEK RIAMREKAKE EK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
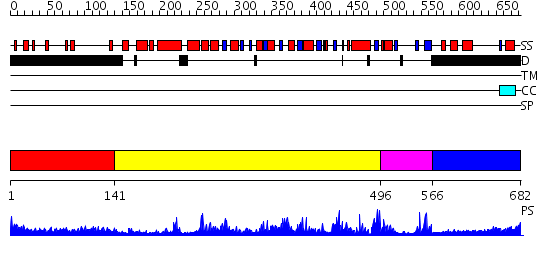
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..140] | 5.268993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [141..495] | 11.522879 | Crystal structure of Tt1595, a putative SAM-dependent methyltransferase from Thermus thermophillus HB8 | |
| 3 | View Details | [496..565] | 88.045757 | Crystal Structure Analysis of Methyltransferase Homolog Protein from Pyrococcus Horikoshii | |
| 4 | View Details | [566..682] | 1.13 | No description for 2j63A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)