













| General Information: |
|
| Name(s) found: |
RH48_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 798 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] ATP-dependent helicase activity [IEA][ISS] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYSLILRERS GSITGSLWNR ISSRNMGGGP RTFPGGLNKW QWKRMHEKKA REKENKLLDQ 60
61 EKQLYEARIR TEIRAKMWGN PDSGEKTAKS KQSHGPMSPK EHIKTLADRF MKAGAEDFWN 120
121 ENDGPVKKSD QGSRSGSDSI DSTSNSPIDV RRLVSATCDS MGKNRVFGSS RRGFSSMSRF 180
181 KRNESSCDEG DDFDAKKLDT LSPFSPKFAG TKEKVKSSRS VVGVIRNKGL FGRRKFRKND 240
241 SSTEEDSDEE GDEGKMIGWM DMRKTGSSAS LGNHDIKLTK RVNRNVTDEE LYPPLDINTV 300
301 REDLSKRKSV DNVMEEKQEP HDSIYSAKRF DESCISPLTL KALSASGILK MTRVQDATLS 360
361 ECLDGKDALV KAKTGTGKSM AFLLPAIETV LKAMNSGKGV NKVAPIFALI LCPTRELASQ 420
421 IAAEGKALLK FHDGIGVQTL IGGTRFKLDQ QRLESEPCQI LIATPGRLLD HIENKSGLTS 480
481 RLMALKLFIV DEADLLLDLG FRRDVEKIID CLPRQRQSLL FSATIPKEVR RVSQLVLKRD 540
541 HSYIDTIGLG CVETHDKVRQ SCIVAPHESH FHLVPHLLKE HINNTPDYKI IVFCSTGMVT 600
601 SLMYTLLREM KLNVREIHAR KPQLHRTRVS DEFKESNRLI LVTSDVSARG MNYPDVTLVI 660
661 QVGIPSDREQ YIHRLGRTGR EGKGGEGLLL IAPWERYFLD ELKDLPLEPI PAPDLDSIVK 720
721 HQVDQSMAKI DTSIKEAAYH AWLGYYNSVR ETGRDKTTLA ELANRFCHSI GLEKPPALFR 780
781 RTAVKMGLKG ISGIPIRK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
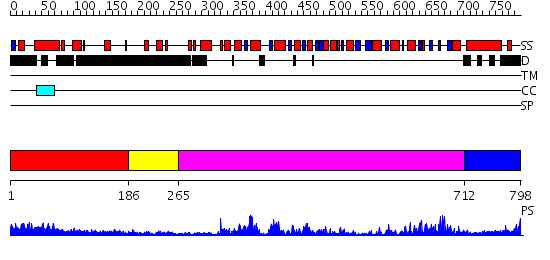
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..185] | 2.263981 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [186..264] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [265..711] | 86.69897 | No description for 2db3A was found. | |
| 4 | View Details | [712..798] | 25.522879 | Nucleotide excision repair enzyme UvrB |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)