













| General Information: |
|
| Name(s) found: |
TAF12_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 539 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
transcription factor TFIID complex [IEA] |
| Biological Process: |
transcription initiation
[IEA]
|
| Molecular Function: |
transcription initiation factor activity
[IEA]
DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDQPRQSSTA SQPPETPPQP SDSKPSTLTQ IQPTPSTNPS PSSVVSSIPS SPAPQSPSLN 60
61 PNPNPPQYTR PVTSPATQQQ QHLSQPLVRP PPQAYSRPWQ QHSSYTHFSS ASSPLLSSSS 120
121 APASSSSSLP ISGQQRGGMA IGVPASPIPS PSPTPSQHSP SAFPGSFGQQ YGGLGRGTVG 180
181 MSEATSNTSS PQVRMMQGTQ GIGMMGTLGS GSQIRPSGMT QHQQRPTQSS LRPASSTSTQ 240
241 SPVAQNFQGH SLMRPSPISS PNVQSTGASQ QSLQAINQPW LSSTPQGKPP LPPPSYRPQV 300
301 NSPSMQQRPH IPQQHISTSA ATPQPQQQQS QQQHQPQEQL QQLRSPQQPL AHPHQPTRVQ 360
361 GLVNQKVTSP VMPSQPPVAQ PGNHAKTVSA ETEPSDDRIL GKRSIHELLQ QIDPSEKLDP 420
421 EVEDILSDIA EDFVESITTF GCSLAKHRKS DILEAKDILL HVERNWNIRP PGFSSDEFKT 480
481 FRKPLTTDIH KERLAAIKKS VTATEAANAR NQFGHGTANA RGGQAKTPSN PMGSTTFNH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
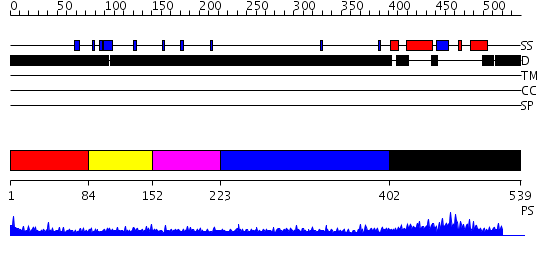
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..83] | 0.991 | Crystal structure of human pregnenolone sulfotransferase (SULT2B1a) in the presence of PAP | |
| 2 | View Details | [84..151] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [152..222] | 1.61 | No description for 2e45A was found. | |
| 4 | View Details | [223..401] | 5.210996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [402..539] | 33.522879 | TAF(II)-20, (TAF(II)-15, hTAF12), histone fold domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)