













| General Information: |
|
| Name(s) found: |
ENO3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 475 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] |
| Biological Process: |
glycolysis
[IEA]
|
| Molecular Function: |
phosphopyruvate hydratase activity
[IEA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVQEYLDKH MLSRKIEDAV NAAVRAKTSD PVLFIANHLK KAVSSVITKV KARQILDSRG 60
61 IPTVEVDLHT NKGVFRASVP SGDSSGTYEA IELRDGDKGM YLGNSVAKAV KNINEKISEA 120
121 LIGMDPKLQG QIDQAMIDLD KTEKKSELGA NAILAVSIAA CKAGAAEKEV PLCKHLSDLS 180
181 GRANMVLPVP AFTVLSGGKH ASNTFAIQEI MILPIGASRF EEALQWGSET YHHLKAVISE 240
241 KNGGLGCNVG EDGGLAPDIS SLKEGLELVK EAINRTGYND KIKIAIDIAA TNFCLGTKYD 300
301 LDIKSPNKSG QNFKSAEDMI DMYKEICNDY PIVSIEDPFD KEDWEHTKYF SSLGICQVVG 360
361 DDLLMSNSKR VERAIQESSC NALLLKVNQI GTVTEAIEVV KMARDAQWGV VTSHRCGETE 420
421 DSFISDLSVG LATGVIKAGA PCRGERTMKY NQLLRIEEEL GDQAVYAGED WKLSL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
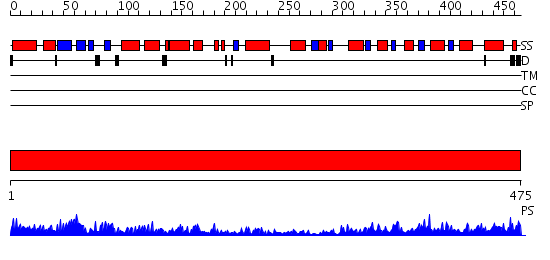
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..475] | 155.0 | Crystal Structure of Human Neuron Specific Enolase at 1.8 angstrom |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)