













| General Information: |
|
| Name(s) found: |
GPDL1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 763 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
apoplast [IDA] anchored to plasma membrane [IDA] anchored to membrane [TAS] |
| Biological Process: |
glycerol metabolic process
[IEA][ISS]
lipid metabolic process [IEA] |
| Molecular Function: |
glycerophosphodiester phosphodiesterase activity
[ISS]
kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSRPSNPTK LVIRSSTLLF CGVVLIHLFA AQIDAQRSTS RWQTLNGDAP LVIARGGFSG 60
61 LYPDSSIAAY QLATLTSVAD VVLWCDLQLT KDGLGICFPD LNLANASTID RVYPNREKSY 120
121 SVNGVTTKGW FPNDFSLTEL QNFLLIRGIL SRTDRFDGNG YLISTIEDVV TTLNREGFWL 180
181 NVQHDAFYEQ QNLSMSSFLL SVSRTVSIDF ISSPEVNFFK KITGSFGRNG PTFVFQFLGK 240
241 EDFEPTTNRT YGSILSNLTF VKTFASGILV PKSYILPLDD EQYLVPHTSL VQDAHKAGLQ 300
301 VYVSGFANDV DIAYNYSSDP VSEYLSFVDN GDFSVDGVLS DFPITASAAV DCFSHIGRNA 360
361 TKQVDFLVIS KDGASGDYPG CTDLAYEKAI KDGADVIDCS VQMSSDGVPF CLRSIDLRNS 420
421 IAALQNTFSN RSTSVPEISS VPGIFTFSLT WPEIQSLTPA ISNPFRVYRI FRNPREKNSG 480
481 KLISLSQFLD LAKTYTSLSG VLISVENAAY LREKQGLDVV QAVLDTLTEA GYSNGTTTTK 540
541 VMIQSTNSSV LVDFKKQSKY ETVYKIEETI GNIRDSAIED IKKFANAVVI NKDSVFPNSD 600
601 SFLTGQTNVV ERLQKSQLPV YVELFRNEFV SQAYDFFSDA TVEINAYIYG AGINGTITEF 660
661 PFTAARYKRN RCLGREEVPP YMLPVNPGGL LNVMSPLSLP PAQAPNQDFI EADVTEPPLS 720
721 PVIAKAPTST PGTPSTIAQA PSGQTRLKLS LLLSVFFLSL LLL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
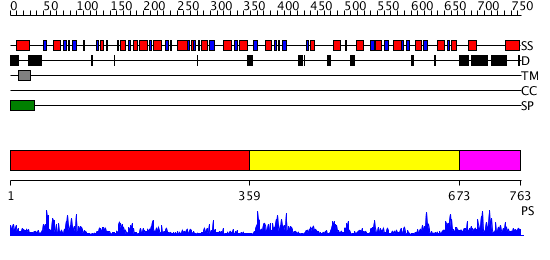
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..358] | 7.78 | Crystal structure of periplasmic glycerophosphodiester phosphodiesterase from Escherichia coli | |
| 2 | View Details | [359..672] | 65.69897 | Structural genomics, Crystal structure of Glycerophosphoryl diester phosphodiesterase from E. coli | |
| 3 | View Details | [673..763] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|