













| General Information: |
|
| Name(s) found: |
DNMT2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1512 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
DNA methylation
[IEA][ISS]
|
| Molecular Function: |
DNA (cytosine-5-)-methyltransferase activity
[IEA][ISS]
DNA binding [IEA] protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METKVGKQKK RSVDSNDDVS KERRPKRAAA CRNFKEKPLR ISDKSETVEA KKEQNVVEEI 60
61 VAIQLTSSLE SNDDPRPNRR LTDFVLHNSD GVPQPVEMLE LGDIFLEGVV LPLGDDKNEE 120
121 KGVRFQSFGR VENWNISGYE DGSPGIWIST ALADYDCRKP ASKYKKIYDY FFEKACACVE 180
181 VFKSLSKNPD TSLDELLAAV ARSMSGSKIF SSGGAIQEFV ISQGEFIYNQ LAGLDETAKN 240
241 HETCFVENSV LVSLRDHESS KIHKALSNVA LRIDESQLVK SDHLVDGAEA EDVRYAKLIQ 300
301 EEEYRISMER SRNKRSSTTS ASNKFYIKIN EHEIANDYPL PSYYKNTKEE TDELLLFEPG 360
361 YEVDTRDLPC RTLHNWALYN SDSRMISLEV LPMRPCAEID VTVFGSGVVA EDDGSGFCLD 420
421 DSESSTSTQS NVHDGMNIFL SQIKEWMIEF GAEMIFVTLR TDMAWYRLGK PSKQYAPWFE 480
481 TVMKTVRVAI SIFNMLMRES RVAKLSYANV IKRLCGLEEN DKAYISSKLL DVERYVVVHG 540
541 QIILQLFEEY PDKDIKRCPF VTGLASKMQD IHHTKWIIKR KKKILQKGKN LNPRAGLAHV 600
601 VTRMKPMQAT TTRLVNRIWG EFYSIYSPEV PSEAIHEVEE EEIEEDEEED ENEEDDIEEE 660
661 AVEVQKSHTP KKSRGNSEDM EIKWNGEILG ETSDGEPLYG RALVGGETVA VGSAVILEVD 720
721 DPDETPAIYF VEFMFESSDQ CKMLHGKLLQ RGSETVIGTA ANERELFLTN ECLTVHLKDI 780
781 KGTVSLDIRS RPWGHQYRKE NLVVDKLDRA RAEERKANGL PTEYYCKSLY SPERGGFFSL 840
841 PRNDIGLGSG FCSSCKIKEE EEERSKTKLN ISKTGVFSNG IEYYNGDFVY VLPNYITKDG 900
901 LKKGTSRRTT LKCGRNVGLK AFVVCQLLDV IVLEESRKAS NASFQVKLTR FYRPEDISEE 960
961 KAYASDIQEL YYSHDTYILP PEALQGKCEV RKKNDMPLCR EYPILDHIFF CEVFYDSSTG1020
1021 YLKQFPANMK LKFSTIKDET LLREKKGKGV ETGTSSGILM KPDEVPKEMR LATLDIFAGC1080
1081 GGLSHGLEKA GVSNTKWAIE YEEPAGHAFK QNHPEATVFV DNCNVILRAI MEKCGDVDDC1140
1141 VSTVEAAELV AKLDENQKST LPLPGQADFI SGGPPCQGFS GMNRFSDGSW SKVQCEMILA1200
1201 FLSFADYFRP KYFLLENVKK FVTYNKGRTF QLTMASLLEI GYQVRFGILE AGTYGVSQPR1260
1261 KRVIIWAASP EEVLPEWPEP MHVFDNPGSK ISLPRGLHYD TVRNTKFGAP FRSITVRDTI1320
1321 GDLPLVENGE SKINKEYRTT PVSWFQKKIR GNMSVLTDHI CKGLNELNLI RCKKIPKRPG1380
1381 ADWRDLPDEN VTLSNGLVEK LRPLALSKTA KNHNEWKGLY GRLDWQGNLP ISITDPQPMG1440
1441 KVGMCFHPEQ DRIITVRECA RSQGFPDSYE FSGTTKHKHR QIGNAVPPPL AFALGRKLKE1500
1501 ALYLKSSLQH QS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
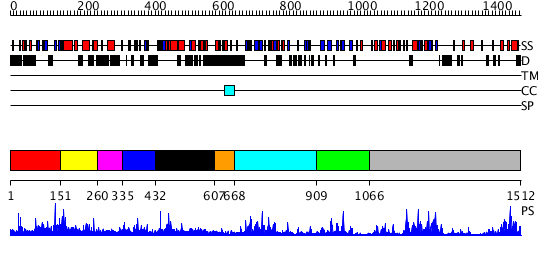
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..150] | 1.011967 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [151..259] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [260..334] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [335..431] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [432..606] | 4.09691 | No description for 2grcA was found. | |
| 6 | View Details | [607..667] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [668..908] | 31.30103 | Crystal structure of the proximal BAH domain of polybromo | |
| 8 | View Details | [909..1065] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1066..1512] | 46.154902 | DNA methylase HaeIII |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)