













| General Information: |
|
| Name(s) found: |
gi|25511671
[NCBI NR]
gi|22022524 [NCBI NR] gi|15217815 [NCBI NR] gi|12323002 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 786 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKERYSRSH RDDRDRDSSP DHSPQREGGR RRDRDVDSKR RDSDHYRSSR RGDREDERDR 60
61 TKDRRGRSVE RGEREGSRDR EKHHHERSHE GSKEKESRSK RKDREEENGA RDGKKKSRFA 120
121 DGNGERRSRF EDVAIEVENK DAQVSEGSGA TNPTSGVTMG ASTYSSIPSE ASAAPSQTLL 180
181 TKVSSISTTD ENKASVVRSH EVPGKSSTDG RPLSTAGKSS ANLPLDSSAL AAKARKALQL 240
241 QKGLADRLKN LPLLKKATKP TSEGSPHTRV PPSTTTPAVS TGTSFASTLP HTGLAGFGSI 300
301 ANIEAVKRAQ ELAANMGFHQ DREFAPVINL FPGQAPSDMT VAQRPEKPPV LRVDALGREI 360
361 DEHGNVISVT KPSNLSTLKV NINKKKKDAF QILKPQLEAD LKENPYFDTR MGIDEKKILR 420
421 PKRMSFQFVE EGKWTRDAEN LKFKSHFGEA KAKELKVKQA QLAKANDDIN PNLIEVSERV 480
481 PRKEKPKEPI PDVEWWDANV LTNGEYGEIT DGTITESHLK IEKLTHYIEH PRPIEPPAEA 540
541 APPPPQPLKL TKKEQKKLRT QRRLAKEKEK QEMIRQGLLE PPKAKVKMSN LMKVLGSEAT 600
601 QDPTKLEKEI RTAAAEREQA HTDRNAARKL TPAEKREKKE RKLFDDPTTV ETIVSVYKIK 660
661 KLSHPKTRFK VEMNARENRL TGCSVMTDEM SVVVVEGKSK AIKRYGKLMM KRINWEEAER 720
721 KEGNEDEEEE VNGGNKCWLV WQGSIGKPSF HRFHVHECVT ESTAKKVFMD AGVVHYWDLA 780
781 VNYSDD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
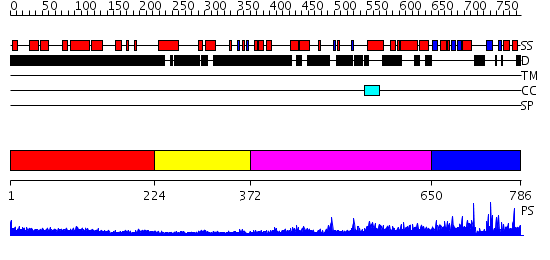
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..223] | 5.182996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [224..371] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [372..649] | 3.30103 | Heavy meromyosin subfragment | |
| 4 | View Details | [650..786] | 1.38 | No description for 2iygA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)