













| General Information: |
|
| Name(s) found: |
ACO32_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 675 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IEA][ISS]
|
| Biological Process: |
fatty acid beta-oxidation
[IEA][ISS]
metabolic process [IEA] fatty acid metabolic process [IEA] |
| Molecular Function: |
acyl-CoA dehydrogenase activity
[IEA]
acyl-CoA oxidase activity [IEA][ISS] oxidoreductase activity, acting on the CH-CH group of donors [IEA] FAD binding [IEA] oxidoreductase activity [IEA] electron carrier activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSENVELRRA HILANHILRS PRPSSNPSLT PEVCFQYSPP ELNESYGFEV KEMRKLLDGH 60
61 NLEERDWLYG LMMQSNLFNP KQRGGQIFVS PDYNQTMEQQ RQISMKRIFY LLEKGVFQGW 120
121 LTETGPEAEL KKFALYEVCG IYDYSLSAKL GVHFLLWGNA VKFFGTKRHH EKWLKDTEDY 180
181 VVKGCFAMTE LGHGTNVRGI ETVTTYDPTT EEFVINTPCE SAQKYWIGEA ANHANHAIVI 240
241 SQLSMNGTNQ GIHVFIAQIR DHDGNTCPNV RIADCGHKIG LNGVDNGRIW FDNLRIPREN 300
301 LLNSVADVLA DGKYVSSIKD PDQRFGAFLA PLTSGRVTIA SSAIYSAKLG LAVAIRYSLS 360
361 RRAFSVAANG PEVLLLDYPS HQRRLLPLLA KTYAMSFAVN DLKMIYVKRT PETNKAIHVV 420
421 SSGFKAVLTW HNMRTLQECR EAVGGQGLKT ENRVGHLKGE YDVQTTFEGD NNVLMQLVSK 480
481 ALFAEYVSCK KRNKPFKGLG LEHMNSPRPV LPTQLTSSTL RCSQFQKSVF CLRERDLLER 540
541 FTSEVAELQG RGESREFLFL LNHQLSEDLS KAFTEKAILQ TVLDAEAKLP PGSVKDVLGL 600
601 VRSMYALISL EEDPSLLRYG HLSRDNVGDV RKEVSKLCGE LRPHALALVA SFGIPDAFLS 660
661 PIAFNWVEAN AWSSL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
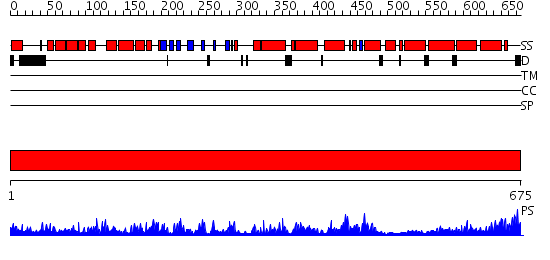
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..675] | 135.0 | Peroxisomal acyl-CoA oxidase-II, domains 3 and 4; Peroxisomal acyl-CoA oxidase-II, domains 1 and 2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)