













| General Information: |
|
| Name(s) found: |
HIS1A_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 411 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
histidine biosynthetic process
[IDA]
|
| Molecular Function: |
ATP phosphoribosyltransferase activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLLLPTNLQ QYPSSSSFPS STPILSPPPS TAFSVIVPRR RCLRLVTSCV STVQSSVATN 60
61 GSSPAPAPAA VVVERDQIRL GLPSKGRMAA DAIDLLKDCQ LFVKQVNPRQ YVAQIPQLPN 120
121 TEVWFQRPKD IVRKLLSGDL DLGIVGLDTL SEYGQENEDL IIVHEALNFG DCHLSIAIPN 180
181 YGIFENINSL KELAQMPQWS EERPLRLATG FTYLGPKFMK ENGIKHVVFS TADGALEAAP 240
241 AMGIADAILD LVSSGITLKE NNLKEIEGGV VLESQAALVA SRRALNERKG ALNTVHEILE 300
301 RLEAHLKADG QFTVVANMRG NSAQEVAERV LSQPSLSGLQ GPTISPVYCT QNGKVSVDYY 360
361 AIVICVPKKA LYDSVKQLRA AGGSGVLVSP LTYIFDEDTP RWGQLLRNLG I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
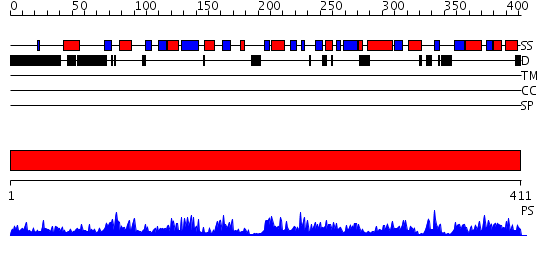
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..411] | 94.69897 | STRUCTURE OF THE E.COLI ATP-PHOSPHORIBOSYLTRANSFERASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)