













| General Information: |
|
| Name(s) found: |
HDG12_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 687 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][ISS]
|
| Biological Process: |
regulation of transcription, DNA-dependent
[ISS]
trichome branching [IGI] |
| Molecular Function: |
transcription factor activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEFLGDSQNH DSSETEKKNK KKKRFHRHTP HQIQRLESTF NECQHPDEKQ RNQLSRELGL 60
61 APRQIKFWFQ NRRTQKKAQH ERADNCALKE ENDKIRCENI AIREAIKHAI CPSCGDSPVN 120
121 EDSYFDEQKL RIENAQLRDE LERVSSIAAK FLGRPISHLP PLLNPMHVSP LELFHTGPSL 180
181 DFDLLPGSCS SMSVPSLPSQ PNLVLSEMDK SLMTNIAVTA MEELLRLLQT NEPLWIKTDG 240
241 CRDVLNLENY ENMFTRSSTS GGKKNNLGME ASRSSGVVFT NAITLVDMLM NSVKLTELFP 300
301 SIVASSKTLA VISSGLRGNH GDALHLMIEE LQVLSPLVTT REFCVLRYCQ QIEHGTWAIV 360
361 NVSYEFPQFI SQSRSYRFPS GCLIQDMSNG YSKVTWVEHG EFEEQEPIHE MFKDIVHKGL 420
421 AFGAERWIAT LQRMCERFTN LLEPATSSLD LGGVIPSPEG KRSIMRLAHR MVSNFCLSVG 480
481 TSNNTRSTVV SGLDEFGIRV TSHKSRHEPN GMVLCAATSF WLPISPQNVF NFLKDERTRP 540
541 QWDVLSNGNS VQEVAHITNG SNPGNCISVL RGFNASSSQN NMLILQESCI DSSSAALVIY 600
601 TPVDLPALNI AMSGQDTSYI PILPSGFAIS PDGSSKGGGS LITVGFQIMV SGLQPAKLNM 660
661 ESMETVNNLI NTTVHQIKTT LNCPSTA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
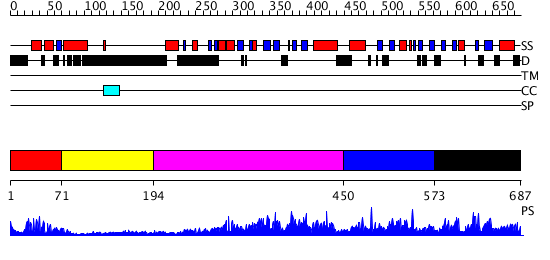
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] | 19.69897 | Paired protein | |
| 2 | View Details | [71..193] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [194..449] | 2.79 | No description for 2r55A was found. | |
| 4 | View Details | [450..572] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [573..687] | 2.055976 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)