













| General Information: |
|
| Name(s) found: |
gi|7547107
[NCBI NR]
gi|15229304 [NCBI NR] |
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 715 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endomembrane system
[IEA]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
endoribonuclease activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKVVALVSGG KDSCYAMMKC IQYGHEIVAL ANLLPVDDSV DELDSYMYQT VGHQILVGYA 60
61 ECMNVPLFRR RIRGSSRHQK LSYQMTPDDE VEDMFVLLSE VKRQIPSITA VSSGAIASDY 120
121 QRLRVESICS RLGLVSLAFL WKQDQTLLLQ DMIANGIKAI LVKVAAIGLD PSKHLGKDLA 180
181 FMEPYLLKLK EKYGSNVCGE GGEYETLTLD CPLFTNASIV LDEYQVVLHS PDSIAPVGVL 240
241 HPSTFHLEKK GNPDSHSPEE ESSLVSEVLG DGPNTSDSTR QRDNGIVDLV EHTSNRLHIS 300
301 RAEKHNTFSI CCWLEDSSES SKEDLETVLT ELESQLLKHG YNWQHVLYIH LYISDMSEFA 360
361 VANETYVKFI TQEKCPFGVP SRSTIELPLV QAGLGKAYIE VLVANDESKR VLHVQSISCW 420
421 APSCIGPYSQ ATLHQSVLHM AGQLGLDPPT MNLQTEGAIA ELNQALTNSE AIAESFNCSI 480
481 SSSAILFVVF CSARTKQSER NQLHEKFVTF LGLAKSSRRV QNVLDPMFLY ILVPDLPKRA 540
541 LVEVKPILYV EEDTDTEDET SRDQSGEGHY SIWGYKPEKW HQDCVQKRVV DGKVCVAVLS 600
601 ISAELMRKLQ GEEEELEIVS RFCVYLLNKT LSENSFSWQD TTSLRIHFST SIGVSVERLS 660
661 AIFVSAFREL NEMSDGVKMD SLKEPIFNLV PVLGAGNSSA SLDNIITCEL FALRS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
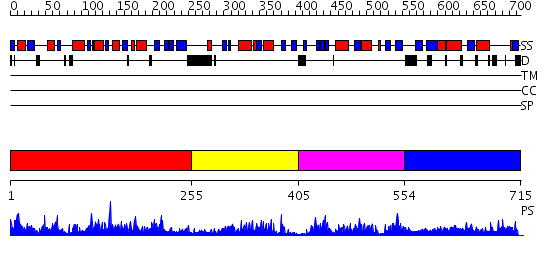
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..254] | 70.39794 | Crystal Structure of the putative n-type ATP pyrophosphatase from Pyrococcus furiosus, the Northeast Structural Genomics Target PfR23 | |
| 2 | View Details | [255..404] | 7.522879 | Structure of hypothetical protein from Streptococcus pyogenes M1 GAS, member of highly conserved yjgF family of proteins | |
| 3 | View Details | [405..553] | 43.0 | No description for 2dyyA was found. | |
| 4 | View Details | [554..715] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 |
|
||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)