













| General Information: |
|
| Name(s) found: |
DOT3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 607 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
phloem or xylem histogenesis
[IMP]
shoot development [IMP] primary root development [IMP] cotyledon vascular tissue pattern formation [IMP] leaf vascular tissue pattern formation [IMP] response to light stimulus [ISS] |
| Molecular Function: |
protein binding
[IEA]
signal transducer activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSVSAAERL KSFGDAGPVC NKSIIFPSRV VTLANSFEKK DRSWYVKSQI PTDLSIQVND 60
61 ITFKAHKFPL ISKCGYISSI ELKPSTSENG YHLKLENFPG GADTFETILK FCYNLPLDLN 120
121 PLNVAPLRCA SEYLYMTEEF EAGNLISKTE AFITFVVLAS WRDTLTVLRS CTNLSPWAEN 180
181 LQIVRRCCDL LAWKACNDNN IPEDVVDRNE RCLYNDIATL DIDHFMRVIT TMKARRAKPQ 240
241 ITGKIIMKYA DNFLPVINDD LEGIKGYGLG KNELQFSVNR GRMEESNSLG CQEHKETIES 300
301 LVSVLPPQSG AVSCHFLLRM LKTSIVYSAS PALISDLEKR VGMALEDANV CDLLIPNFKN 360
361 EEQQERVRIF EFFLMHEQQQ VLGKPSISKL LDNYLAEIAK DPYLPITKFQ VLAEMLPENA 420
421 WKCHDGLYRA IDMFLKTHPS LSDHDRRRLC KTMNCEKLSL DACLHAAQND RLPLRTIVQI 480
481 NTQVLFSEQV KMRMMMQDKL PEKEEENSGG REDKRMSRDN EIIKTLKEEL ENVKKKMSEL 540
541 QSDYNELQQE YERLSSKQKS SHNWGLRWQK VKKSFQTKRE DEETRERTRR RSSTGQRTSF 600
601 RRRMSMS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
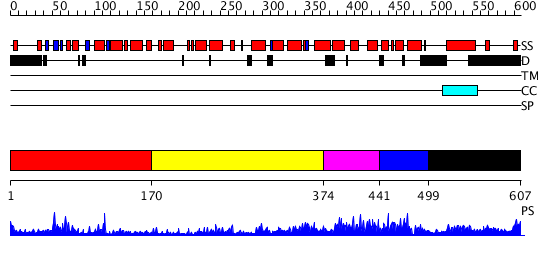
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..169] | 16.045757 | No description for 2ihcA was found. | |
| 2 | View Details | [170..373] | 4.624986 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [374..440] | 1.193985 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [441..498] | 2.074989 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [499..607] | 1.071984 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)