













| General Information: |
|
| Name(s) found: |
TRP4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 640 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
chromosome, telomeric region [IDA] |
| Biological Process: |
telomere maintenance
[TAS]
|
| Molecular Function: |
DNA binding
[IDA][ISS]
double-stranded telomeric DNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVKRKLNCG GSNGFDFPNI PKAPRSSRRK VSGKRSDDES EICAIDLLAS LAGKLLEESE 60
61 SSSTSTYASE ADNLDHLGGL IKQELEDGYT TKPCKSEFFD PGNPASKSTS ENTSVTCLPF 120
121 SSFENDCILE QTPVSDCKRA SGLKSLVGSI TEETCVVNED AGSEQGANTF SLKDPSQLHS 180
181 QSPESVLLDG DVKLAPCTDQ VPNDSFKGYR NHSKLVCRDD DENYCKYYKF SDKCKSYRPL 240
241 SRVGNRRIMQ SVRAISKLKC FEDTRTDGRL KALYRKRKLC YGYNPWKRET IHRKRRLSDK 300
301 GLVVNYDGGL SSESVSNSPE KGESENGDFS AAKIGLLSKD SRVKFSIKSL RIPELVIEVP 360
361 ETATVGLLKR TVKEAVTALL GGGIRIGVLV QGKKVRDDNN TLSQTGLSCR ENLGNLGFTL 420
421 EPGLETLPVP LCSETPVLSL PTDSTKLSER SAASPALETG IPLPPQDEDY LINLGNSVEN 480
481 NDELVPHLSD IPADEQPSSD SRALVPVLAL ESDALALVPV NEKPKRTELS QRRTRRPFSV 540
541 TEVEALVSAV EEVGTGRWRD VKLRSFENAS HRTYVDLKDK WKTLVHTASI SPQQRRGEPV 600
601 PQELLDRVLG AHRYWTQHQM KQNGKHQVAT TMVVEAGSSM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
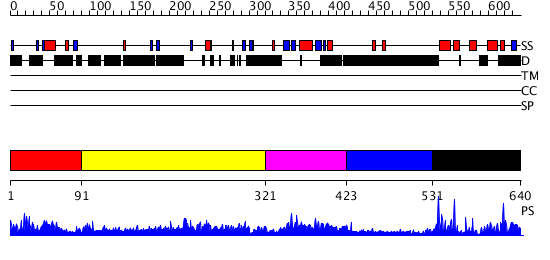
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [91..320] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [321..422] | 1.12 | Solution Structure of the Ubiquitin-like Domain from Mouse Hypothetical 1700011N24Rik Protein | |
| 4 | View Details | [423..530] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [531..640] | 14.69897 | No description for 2ajeA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)