













| General Information: |
|
| Name(s) found: |
gi|8809580
[NCBI NR]
gi|15239546 [NCBI NR] |
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 795 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
maintenance of fidelity involved in DNA-dependent DNA replication
[IEA]
mismatch repair [IEA][ISS] |
| Molecular Function: |
ATP binding
[IEA][ISS]
mismatched DNA binding [IEA] damaged DNA binding [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQTLSIAFCN SALVIRRRNS IGNRNRVKLS LISSSSPTLV CHSKSKSQTD SLRVLEWDKL 60
61 CDVVASFART SLGREATKKK LWSLDQSFSE SLKLLDETDA AIKMLEHGSF CLDLSSIHIS 120
121 LVESGIRHAK RRLSLRADQA LEVASLLRFF ENLQLDLKAA IKQDGDWYKR FMPLSELIVH 180
181 PVINRSFVKL VEQVIDPDGT IKDSASSALR QSRERVQTLE RKLQQLLDAI IRSQKDDESV 240
241 MIKFQLAAEI DGRWCIQMSS NQLTSVNGLL LSSVYFQVLS LEDNMCFSGS GGGTAAEPIA 300
301 AVSMNDDLQS ARASVAKAEA EILSMLTEKI NARATYSRAY GGAHPDIYLP PEDEVESLSA 360
361 GENSPDINLP SEKPLSKKEW LLYLPRCYHP LLLYQHKKGI RKTRETVKFH KTADTVLSGA 420
421 PPIPADFQIS KGTRVLVITG PNTGGKTICL KSVGLAAMMA KSGLYVLATE SARIPWFDNI 480
481 YADIGDEQSL LQSLSTFSGH LKQISEILSH STSRSLVLLD EVGAGTNPLE GAALGMAILE 540
541 SFAESGSLLT MATTHHGELK TLKYSNSAFE NACMEFDDLN LKPTYKILWG VPGRSNAINI 600
601 ADRLGLPCDI IESARELYGS ASAEINEVIL DMERYKQEYQ RLLNESRVYI RLSRELHENL 660
661 LTAQKNINDH STKERRKMRQ ELTQAGSMTR STLRRTLQQF RSSAGKSSQS KVATQLQTKV 720
721 KTTKDEDNGI RSSSVVERRP LPEAAAQKVP EVGSSVFVSS LGKKATVLKV EHSKKEILVQ 780
781 VGIMKMKVKL TDVVA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
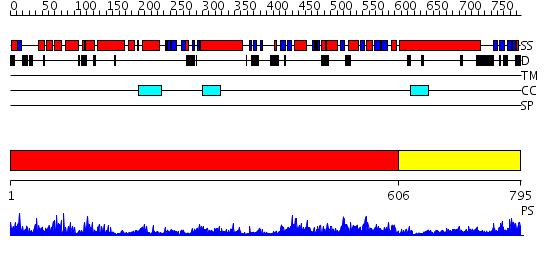
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..605] | 137.0 | Crystal Structure of E. coli DNA Mismatch Repair enzyme MutS, E38T mutant, in complex with a G.T mismatch | |
| 2 | View Details | [606..795] | 82.09691 | Crystal Structure Analysis of ClpB |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.64 |
Source: Reynolds et al. (2008)