













| General Information: |
|
| Name(s) found: |
FHY3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 839 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
red, far-red light phototransduction
[IMP]
response to far red light [IMP] red or far-red light signaling pathway [IMP] circadian rhythm [IMP] |
| Molecular Function: |
transcription factor activity
[IDA]
transcription activator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDIDLRLHSG DLCKGDDEDR GLDNVLHNEE DMDIGKIEDV SVEVNTDDSV GMGVPTGELV 60
61 EYTEGMNLEP LNGMEFESHG EAYSFYQEYS RAMGFNTAIQ NSRRSKTTRE FIDAKFACSR 120
121 YGTKREYDKS FNRPRARQSK QDPENMAGRR TCAKTDCKAS MHVKRRPDGK WVIHSFVREH 180
181 NHELLPAQAV SEQTRKIYAA MAKQFAEYKT VISLKSDSKS SFEKGRTLSV ETGDFKILLD 240
241 FLSRMQSLNS NFFYAVDLGD DQRVKNVFWV DAKSRHNYGS FCDVVSLDTT YVRNKYKMPL 300
301 AIFVGVNQHY QYMVLGCALI SDESAATYSW LMETWLRAIG GQAPKVLITE LDVVMNSIVP 360
361 EIFPNTRHCL FLWHVLMKVS ENLGQVVKQH DNFMPKFEKC IYKSGKDEDF ARKWYKNLAR 420
421 FGLKDDQWMI SLYEDRKKWA PTYMTDVLLA GMSTSQRADS INAFFDKYMH KKTSVQEFVK 480
481 VYDTVLQDRC EEEAKADSEM WNKQPAMKSP SPFEKSVSEV YTPAVFKKFQ IEVLGAIACS 540
541 PREENRDATC STFRVQDFEN NQDFMVTWNQ TKAEVSCICR LFEYKGYLCR HTLNVLQCCH 600
601 LSSIPSQYIL KRWTKDAKSR HFSGEPQQLQ TRLLRYNDLC ERALKLNEEA SLSQESYNIA 660
661 FLAIEGAIGN CAGINTSGRS LPDVVTSPTQ GLISVEEDNH SRSAGKTSKK KNPTKKRKVN 720
721 PEQDVMPVAA PESLQQMDKL SPRTVGIESY YGTQQSVQGM VQLNLMGPTR DNFYGNQQTM 780
781 QGLRQLNSIA PSYDSYYGPQ QGIHGQGVDF FRPANFSYDI RDDPNVRTTQ LHEDASRHS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
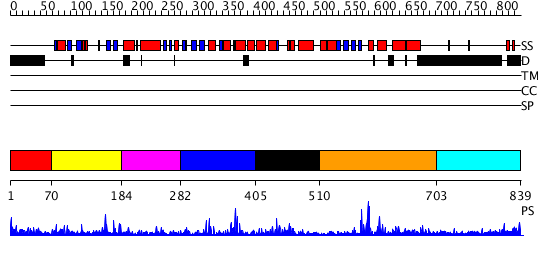
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..69] | 2.371995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [70..183] | 31.207608 | No description for PF03101.7 was found. No confident structure predictions are available. | |
| 3 | View Details | [184..281] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [282..404] | 28.443697 | No description for PF10551.1 was found. No confident structure predictions are available. | |
| 5 | View Details | [405..509] | 8.699997 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [510..702] | 6.486989 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [703..839] | 1.139995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)