













| General Information: |
|
| Name(s) found: |
FRS8_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 696 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to red or far red light
[ISS]
|
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEEQLVVDDD DDDLAPPPIP DLDIDLEDDD DACCHGLLHI APNHEEETGC DENAFANEKC 60
61 LMAPPPTPGM EFESYDDAYS FYNSYARELG FAIRVKSSWT KRNSKEKRGA VLCCNCQGFK 120
121 LLKDAHSRRK ETRTGCQAMI RLRLIHFDRW KVDQVKLDHN HSFDPQRAHN SKSHKKSSSS 180
181 ASPATKTNPE PPPHVQVRTI KLYRTLALDT PPALGTSLSS GETSDLSLDH FQSSRRLELR 240
241 GGFRALQDFF FQIQLSSPNF LYLMDLADDG SLRNVFWIDA RARAAYSHFG DVLLFDTTCL 300
301 SNAYELPLVA FVGINHHGDT ILLGCGLLAD QSFETYVWLF RAWLTCMLGR PPQIFITEQC 360
361 KAMRTAVSEV FPRAHHRLSL THVLHNICQS VVQLQDSDLF PMALNRVVYG CLKVEEFETA 420
421 WEEMIIRFGM TNNETIRDMF QDRELWAPVY LKDTFLAGAL TFPLGNVAAP FIFSGYVHEN 480
481 TSLREFLEGY ESFLDKKYTR EALCDSESLK LIPKLKTTHP YESQMAKVFT MEIFRRFQDE 540
541 VSAMSSCFGV TQVHSNGSAS SYVVKEREGD KVRDFEVIYE TSAAAQVRCF CVCGGFSFNG 600
601 YQCRHVLLLL SHNGLQEVPP QYILQRWRKD VKRLYVAEFG SGRVDIMNPD QWYEHLHRRA 660
661 MQVVEQGMRS KEHCRAAWEA FRECANKVQF VTEKPS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
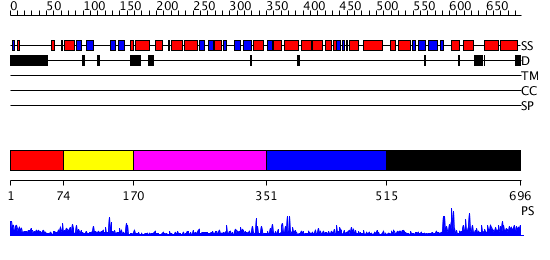
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..73] | 1.357991 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [74..169] | 35.958607 | No description for PF03101.6 was found. No confident structure predictions are available. | |
| 3 | View Details | [170..350] | 8.683995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [351..514] | 20.703996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [515..696] | 5.534996 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)