













| General Information: |
|
| Name(s) found: |
GAI_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 533 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS][TAS]
|
| Biological Process: |
hyperosmotic salinity response
[IGI]
negative regulation of seed germination [IGI] response to salt stress [IGI] regulation of oxygen and reactive oxygen species metabolic process [IGI] salicylic acid mediated signaling pathway [IGI] gibberellic acid mediated signaling pathway [TAS] jasmonic acid mediated signaling pathway [IGI] response to ethylene stimulus [IGI] negative regulation of gibberellic acid mediated signaling pathway [IMP][TAS] response to gibberellin stimulus [IEP] response to abscisic acid stimulus [IGI] response to far red light [IEP] regulation of nitrogen utilization [IMP] phloem transport [IMP] |
| Molecular Function: |
transcription factor activity
[ISS][TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRDHHHHHH QDKKTMMMNE EDDGNGMDEL LAVLGYKVRS SEMADVAQKL EQLEVMMSNV 60
61 QEDDLSQLAT ETVHYNPAEL YTWLDSMLTD LNPPSSNAEY DLKAIPGDAI LNQFAIDSAS 120
121 SSNQGGGGDT YTTNKRLKCS NGVVETTTAT AESTRHVVLV DSQENGVRLV HALLACAEAV 180
181 QKENLTVAEA LVKQIGFLAV SQIGAMRKVA TYFAEALARR IYRLSPSQSP IDHSLSDTLQ 240
241 MHFYETCPYL KFAHFTANQA ILEAFQGKKR VHVIDFSMSQ GLQWPALMQA LALRPGGPPV 300
301 FRLTGIGPPA PDNFDYLHEV GCKLAHLAEA IHVEFEYRGF VANTLADLDA SMLELRPSEI 360
361 ESVAVNSVFE LHKLLGRPGA IDKVLGVVNQ IKPEIFTVVE QESNHNSPIF LDRFTESLHY 420
421 YSTLFDSLEG VPSGQDKVMS EVYLGKQICN VVACDGPDRV ERHETLSQWR NRFGSAGFAA 480
481 AHIGSNAFKQ ASMLLALFNG GEGYRVEESD GCLMLGWHTR PLIATSAWKL STN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
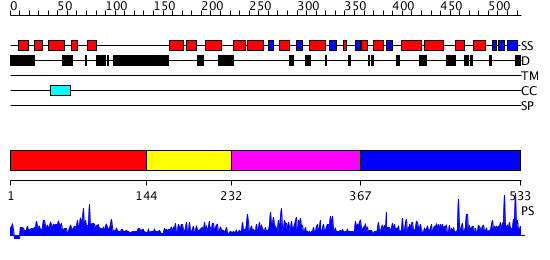
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | 3.057975 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [144..231] | 3.121994 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [232..366] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [367..533] | 5.130963 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)