













| General Information: |
|
| Name(s) found: |
AB6I_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 338 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] |
| Biological Process: |
embryonic development ending in seed dormancy
[IMP]
thylakoid membrane organization [IMP] iron-sulfur cluster assembly [IGI] |
| Molecular Function: |
ATPase activity, coupled to transmembrane movement of substances
[IDA]
protein binding [IPI] transporter activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGVNLQLRH AYSIAQFVPT VSSPPPLPTQ RVRLGTSPSR VLLCNLRANS AAAPILRTTR 60
61 RSVIVSASSV SSAVDSDSLV EDRDDVGRIP LLEVRDLRAV IAESRQEILK GVNLVVYEGE 120
121 VHAVMGKNGS GKSTFSKVLV GHPDYEVTGG SIVFKGQNLL DMEPEDRSLA GLFMSFQSPV 180
181 EIPGVSNMDF LNMAFNARKR KLGQPELDPI QFYSHLVSKL EVVNMKTDFL NRNVNEGFSG 240
241 GERKRNEILQ LAVLGAELAI LDEIDSGLDV DALQDVAKAV NGLLTPKNSV LMITHYQRLL 300
301 DYIKPTLIHI MENGRIIKTG DNSLAKLLEK EGYKAISG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
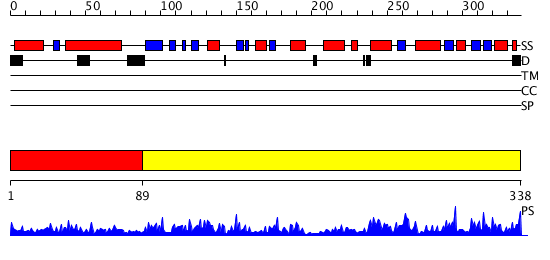
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [89..338] | 44.0 | Crystal Structure of Escherichia coli SufC, an ATPase compenent of the SUF iron-sulfur cluster assembly machinery |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)