













| General Information: |
|
| Name(s) found: |
gi|79313139
[NCBI NR]
gi|6437559 [NCBI NR] gi|15230701 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 673 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein folding
[IEA][ISS]
|
| Molecular Function: |
heat shock protein binding
[IEA]
unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSINRDEALR AKDLAEGLMK KTDFTAARKL AMKAQKMDSS LENISRMIMV CDVHCAATEK 60
61 LFGTEMDWYG ILQVEQIAND VIIKKQYKRL ALLLHPDKNK LPGAESAFKL IGEAQRILLD 120
121 REKRTLHDNK RKTWRKPAAP PYKAQQMPNY HTQPHFRASV NTRNIFTELR PEIRHPFQKA 180
181 QAQPAAFTHL KTFGTSCVFC RVRYEYDRAH VNKEVTCETC KKRFTAFEEP LQSAPQAKGP 240
241 SQTTYCFPQQ SKFPDQRACS EPHKRPENPP TVSSSKASFP MPGSTAKHNG KRKRKNVAEC 300
301 SESSDSESSS ESEDDVNNDT TAAQDSGSNG GEQPRRSVRS KQKVSYNENL SDDDVDLVND 360
361 NGEGSGKNID TEREKETEEE KQTNENHSST ESIDMNGKIE VDQVETPSGA SDSEEDLSSG 420
421 SAEKPNLINY DDPDFNDFDK LREKSCFQAG QIWAVYDEEE GMPRFYALIK KVTTPDFMLR 480
481 YVWFEVDQDQ ENETPNLPVS VGKFVVGNIE ETNLCSIFSH FVYSTTKIRT RKFTVFPKKG 540
541 EIWALFKNWD INCSADSVSP MKYEYEFVEI LSDHAEGATV SVGFLSKVQG FNCVFCPMPK 600
601 DESNTCEIPP HEFCRFSHSI PSFRLTGTEG RGITKGWYEL DPAALPASVS QNLSGEEAAQ 660
661 DRDRQSPPSG SAS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
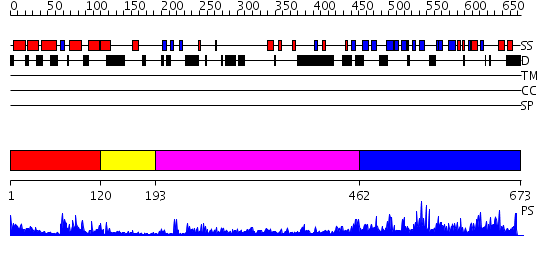
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..119] | 3.69897 | Crystal structure of an 8 repeat consensus TPR superhelix (trigonal crystal form) | |
| 2 | View Details | [120..192] | 22.09691 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 3 | View Details | [193..461] | 22.522879 | The crystal structure of Hsp40 Ydj1 | |
| 4 | View Details | [462..673] | 2.089948 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 |
|
||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)