













| General Information: |
|
| Name(s) found: |
TPPG_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 377 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
metabolic process
[IEA]
trehalose biosynthetic process [IEA][ISS] |
| Molecular Function: |
catalytic activity
[IEA]
trehalose-phosphatase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLNINKTTP VLSDPTTPVS KTRLGSSFPS GRFMMNSRKK IPKLDDVRSN GWLDAMISSS 60
61 PPRKRLVKDF NIEIAPEDDF SQRAWMLKYP SAITSFAHIA AQAKNKKIAV FLDYDGTLSP 120
121 IVDDPDRAIM SDAMRAAVKD VAKYFPTAII SGRSRDKVYQ LVGLTELYYA GSHGMDIMTP 180
181 VNPNGSPEDP NCIKTTDQQG EEVNLFQPAK EFIPVIEEVY NNLVEITKCI KGAKVENHKF 240
241 CTSVHYRNVD EKDWPLVAQR VHDHLKRYPR LRITHGRKVL EVRPVIEWNK GKAVEFLLES 300
301 LGLSNNDEFL PIFIGDDKTD EDAFKVLREG NRGFGILVSS VPKESNAFYS LRDPSEVKKF 360
361 LKTLVKWGKM ESSKTSF |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
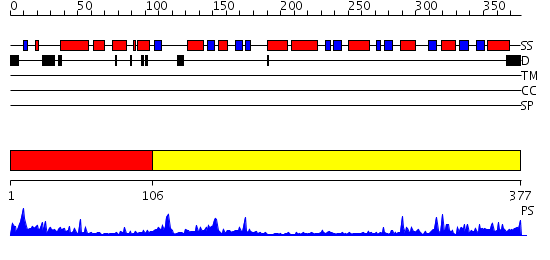
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..105] | 7.39794 | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. | |
| 2 | View Details | [106..377] | 37.69897 | Crystal structure of NYSGRC target T1436: A Hypothetical protein yidA. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)