













| General Information: |
|
| Name(s) found: |
UBP21_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 732 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
ubiquitin-dependent protein catabolic process
[IEA]
|
| Molecular Function: |
ubiquitin-specific protease activity
[ISS]
ubiquitin thiolesterase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEFSDPPPS NLSSSHKLTK PNQTLDESSP TAPIRDLVTN SLSLSSPIRQ IQALSPAKPD 60
61 GSSSSPPDKT LNFNPEENRD VNPDESSSSP SDKTLIAPPA QISPVSNNNH LRITNTSDSY 120
121 LYRPPRRYIE YESDDDELNK MEPTKPLQLS WWYPRIEPTG VGAGLYNSGN TCFIASVLQC 180
181 FTHTVPLIDS LRSFMYGNPC NCGNEKFCVM QALRDHIELA LRSSGYGINI DRFRDNLTYF 240
241 SSDFMINHQE DAHEFLQSFL DKLERCCLDP KNQLGSVSSQ DLNIVDNVFG GGLMSTLCCC 300
301 NCNSVSNTFE PSLGWSLEIE DVNTLWKALE SFTCVEKLED QLTCDNCKEK VTKEKQLRFD 360
361 KLPPVATFHL KRFTNDGVTM EKIFDHIEFP LELDLSPFMS SNHDPEVSTR YHLYAFVEHI 420
421 GIRATFGHYS SYVRSAPETW HNFDDSKVTR ISEERVLSRP AYILFYAREG TPWFSSTFEQ 480
481 LKTVFEATPL HFSPVSVLDN SYESVDNSSK ACNDSVGVSI PDVKWPDSCC QEPKEEVFHS 540
541 AESSNNEDSS AMIDALGSPQ SEKPFAETSQ QTEPESCPTE NKAYIDKSEK PFAETSQPKE 600
601 PKPFADRASI DAPLLKVQNQ DISPKRKAGE RATLGGPKLK YQKPNSHQKR QGTFQIQRAH 660
661 LQTKKQEESR KTKRPLFRSN VAASAPDPKY KNHALSYLNR AQTPRARKLA NALSDSPTKK 720
721 KKSSNMRRSI KL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
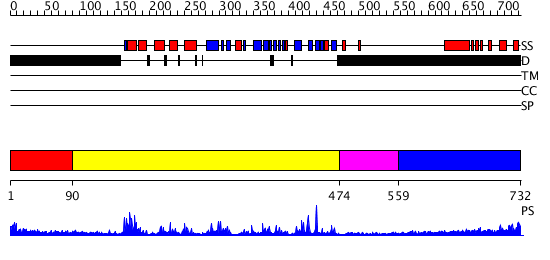
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [90..473] | 61.522879 | No description for 2gfoA was found. | |
| 3 | View Details | [474..558] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [559..732] | 7.284997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)