













| General Information: |
|
| Name(s) found: |
HDG9_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 718 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][ISS]
|
| Biological Process: |
regulation of transcription, DNA-dependent
[IEA][ISS]
regulation of transcription [IEA] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] sequence-specific DNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDFTRDDNSS DERENDVDAN TNNRHEKKGY HRHTNEQIHR LETYFKECPH PDEFQRRLLG 60
61 EELNLKPKQI KFWFQNKRTQ AKSHNEKADN AALRAENIKI RRENESMEDA LNNVVCPPCG 120
121 GRGPGREDQL RHLQKLRAQN AYLKDEYERV SNYLKQYGGH SMHNVEATPY LHGPSNHAST 180
181 SKNRPALYGT SSNRLPEPSS IFRGPYTRGN MNTTAPPQPR KPLEMQNFQP LSQLEKIAML 240
241 EAAEKAVSEV LSLIQMDDTM WKKSSIDDRL VIDPGLYEKY FTKTNTNGRP ESSKDVVVVQ 300
301 MDAGNLIDIF LTAEKWARLF PTIVNEAKTI HVLDSVDHRG KTFSRVIYEQ LHILSPLVPP 360
361 REFMILRTCQ QIEDNVWMIA DVSCHLPNIE FDLSFPICTK RPSGVLIQAL PHGFSKVTWI 420
421 EHVVVNDNRV RPHKLYRDLL YGGFGYGARR WTVTLERTCE RLIFSTSVPA LPNNDNPGVV 480
481 QTIRGRNSVM HLGERMLRNF AWMMKMVNKL DFSPQSETNN SGIRIGVRIN NEAGQPPGLI 540
541 VCAGSSLSLP LPPVQVYDFL KNLEVRHQWD VLCHGNPATE AARFVTGSNP RNTVSFLEPS 600
601 IRDINTKLMI LQDSFKDALG GMVAYAPMDL NTACAAISGD IDPTTIPILP SGFMISRDGR 660
661 PSEGEAEGGS YTLLTVAFQI LVSGPSYSPD TNLEVSATTV NTLISSTVQR IKAMLKCE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
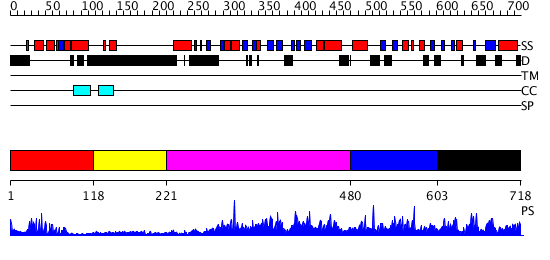
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | 18.39794 | Paired protein | |
| 2 | View Details | [118..220] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [221..479] | 3.68 | No description for 2r55A was found. | |
| 4 | View Details | [480..602] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [603..718] | 2.05598 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)