













| General Information: |
|
| Name(s) found: |
ORP1A_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 760 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
steroid metabolic process
[IEA][ISS]
|
| Molecular Function: |
oxysterol binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYAATPETPF GSARSQPVIT RSVSQRYNHP GQSNHHHLLH SLSFNHQNVL ALPAAAREPP 60
61 VDVKINDIAG NSIAGILYKW VNYGKGWRPR WFVLQDGVLS YYKIKGPDKI VVIHETEKGS 120
121 RVIGEESTRM ISRNKRHAAT NNTNHQLRRK PFGEVHLKVS SIRESRSDDK RFSIFTGTKR 180
181 LHLRAETRED REAWIEALQA VKDMFPRMSN CELMAPTNNL DISIEKLRLR LVEEGVSESA 240
241 IQDCEQITRS EFSAIQSQLL LLKQKQWLLI DTLRQLETEK VDLENTVVDE TQRQAGNGDS 300
301 EETISESDDD NEQFDEAEEE MDTCDSLSSS SFKSIGSVFR TSSFSSDDDG LTNGFESEND 360
361 DVDPSIKTIG FNYPHVKRRK KLPDPVEKEK SVSLWSMIKD NIGKDLTKVC LPVYFNEPLS 420
421 SLQKCFEDLE YSYLLDQASE WGKRGNNLMR ILNVAAFAVS GYASTEGRIC KPFNPMLGET 480
481 YEADYPDKGL RFFSEKVSHH PMIVACHCDG TGWKFWGDSN LKSKFWGRSI QLDPIGLLTL 540
541 QFDDGEIVQW SKVTTSIYNL ILGKLYCDHY GTMKIEGNGE YSCKLKFKEQ SMIDRNPHQV 600
601 QGIVEDKNGK TVARLFGKWD ESIHYVMVDQ GKVNESHLLW KRNKQPENPT KYNLTRFGIT 660
661 LNELTPGLKE KLPPTDSRLR PDQRYLEKGE YEMGNAEKLR LEQRQRQARE MQERGWKPKW 720
721 FRKEKGSETY RYIGGYWEAR DSGSWDDCPD IFGQVHQSIK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
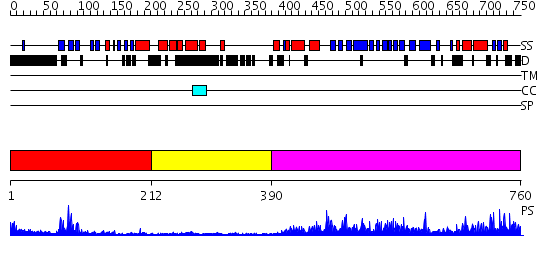
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..211] | 15.09691 | Grp1 | |
| 2 | View Details | [212..389] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [390..760] | 65.221849 | Structure of yeast oxysterol binding protein Osh4 in complex with 7-hydroxycholesterol |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)