













| General Information: |
|
| Name(s) found: |
gi|15230755
[NCBI NR]
gi|12322676 [NCBI NR] |
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 763 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
signal transduction
[IEA]
regulation of transcription, DNA-dependent [IEA] protein amino acid phosphorylation [IEA] |
| Molecular Function: |
protein serine/threonine/tyrosine kinase activity
[ISS]
kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKLLQDPIT PNKELLKKMI ELEKSQEHLM QEMSRLKVST ELRKESRIQC SMNLRPSPWK 60
61 FTHKQYLNIL QSMAQSVHAF DLNMRIIFWN AMAEKVYGYS AAEAVGQNPI DVMVDDRDAP 120
121 FAMTIAQLCS NGESWTGKFP VKRRTGEKFS AVTTCSPFYA DDGSLIGIVS ITSDVAPYLN 180
181 PTISLAKLKA SEVETSSTPA RNSFAFKLGL DTKGAVVSKL GLDSDQPIQV AIASKISDLA 240
241 SKVRNKVRSK MPAGDSSVTV FEGETGDSHH SDHGVFGATL SDHMDDAASS GASTPRGDFI 300
301 QSPFGVFTCN DDKFSSEPFI DSSDGYPITL FTSKAEEWMV KKGLSWPWKG NEQEGSRVKP 360
361 TYSVWPCVQN EQKKDKSHQI NRYSGVKSKS HASESNKPTN NKASGLRSSC INANSAISRG 420
421 IISHSTMNKV DTNSNCLEYE ILWDDLTIGE QIGQGSCGTV YHGLWFGSDV AVKLISKQEY 480
481 SEEVIQSFRQ EVSLMQRLRH PNVLLFMGAV TLPQGLCIVS EFLPRGSLFR LLQRNMSKLD 540
541 WRRRINMALD IARGMNYLHR CSPPIIHRDL KSSNLLVDKN LTVKVADFGL SRIKHHTYLT 600
601 SKSGKGMPQW MAPEVLRNES ADEKSDIYSF GVVLWELATE KIPWENLNSM QVIGAVGFMN 660
661 QRLEIPKDID PDWISLIESC WHRDAKLRPT FQELMERLRD LQRKYTIQFQ ATRAALSDKN 720
721 KEREKERVGL FEQQQLVETC CAGVPMSSTI EQGYGIENIM KVG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
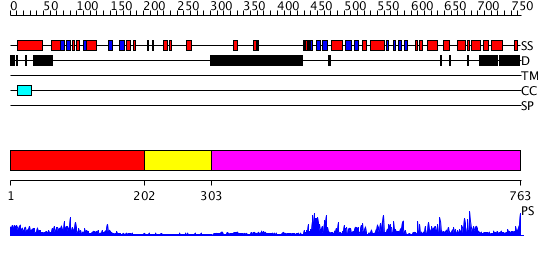
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..201] | 13.221849 | No description for 2gj3A was found. | |
| 2 | View Details | [202..302] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [303..763] | 86.39794 | No description for 2h8hA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.69 |
Source: Reynolds et al. (2008)