













| General Information: |
|
| Name(s) found: |
SUVH9_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 650 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
chloroplast [IEA] |
| Biological Process: |
DNA mediated transformation
[IMP]
|
| Molecular Function: |
zinc ion binding
[IEA]
histone-lysine N-methyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSSHIPLDP SLNPSPSLIP KLEPVTESTQ NLAFQLPNTN PQALISSAVS DFNEATDFSS 60
61 DYNTVAESAR SAFAQRLQRH DDVAVLDSLT GAIVPVEENP EPEPNPYSTS DSSPSVATQR 120
121 PRPQPRSSEL VRITDVGPES ERQFREHVRK TRMIYDSLRM FLMMEEAKRN GVGGRRARAD 180
181 GKAGKAGSMM RDCMLWMNRD KRIVGSIPGV QVGDIFFFRF ELCVMGLHGH PQSGIDFLTG 240
241 SLSSNGEPIA TSVIVSGGYE DDDDQGDVIM YTGQGGQDRL GRQAEHQRLE GGNLAMERSM 300
301 YYGIEVRVIR GLKYENEVSS RVYVYDGLFR IVDSWFDVGK SGFGVFKYRL ERIEGQAEMG 360
361 SSVLKFARTL KTNPLSVRPR GYINFDISNG KENVPVYLFN DIDSDQEPLY YEYLAQTSFP 420
421 PGLFVQQSGN ASGCDCVNGC GSGCLCEAKN SGEIAYDYNG TLIRQKPLIH ECGSACQCPP 480
481 SCRNRVTQKG LRNRLEVFRS LETGWGVRSL DVLHAGAFIC EYAGVALTRE QANILTMNGD 540
541 TLVYPARFSS ARWEDWGDLS QVLADFERPS YPDIPPVDFA MDVSKMRNVA CYISHSTDPN 600
601 VIVQFVLHDH NSLMFPRVML FAAENIPPMT ELSLDYGVVD DWNAKLAICN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
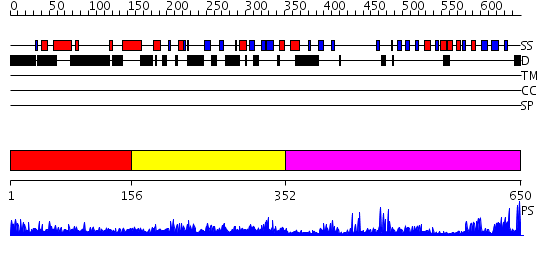
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..155] | 59.109997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [156..351] | 79.958607 | No description for PF02182.8 was found. No confident structure predictions are available. | |
| 3 | View Details | [352..650] | 63.154902 | No description for 2o8jA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)