













| General Information: |
|
| Name(s) found: |
SUVH7_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 693 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
chromatin modification
[IEA]
|
| Molecular Function: |
DNA binding
[IEA]
zinc ion binding [IEA] histone-lysine N-methyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKSIPIKAI PVACVRPDLV DDVTKNTSTI PTMVSPVLTN MPSATSPLLM VPPLRTIWPS 60
61 NKEWYDGDAG PSSTGPIKRE ASDNTNDTAH NTFAPPPEMV IPLITIRPSD DSSNYSCDAG 120
121 AGPSTGPVKR GRGRPKGSKN STPTEPKKPK VYDPNSLKVT SRGNFDSEIT EAETETGNQE 180
181 IVDSVMMRFD AVRRRLCQIN HPEDILTTAS GNCTKMGVKT NTRRRIGAVP GIHVGDIFYY 240
241 WGEMCLVGLH KSNYGGIDFF TAAESAVEGH AAMCVVTAGQ YDGETEGLDT LIYSGQGGTD 300
301 VYGNARDQEM KGGNLALEAS VSKGNDVRVV RGVIHPHENN QKIYIYDGMY LVSKFWTVTG 360
361 KSGFKEFRFK LVRKPNQPPA YAIWKTVENL RNHDLIDSRQ GFILEDLSFG AELLRVPLVN 420
421 EVDEDDKTIP EDFDYIPSQC HSGMMTHEFH FDRQSLGCQN CRHQPCMHQN CTCVQRNGDL 480
481 LPYHNNILVC RKPLIYECGG SCPCPDHCPT RLVQTGLKLH LEVFKTRNCG WGLRSWDPIR 540
541 AGTFICEFAG LRKTKEEVEE DDDYLFDTSK IYQRFRWNYE PELLLEDSWE QVSEFINLPT 600
601 QVLISAKEKG NVGRFMNHSC SPNVFWQPIE YENRGDVYLL IGLFAMKHIP PMTELTYDYG 660
661 VSCVERSEED EVLLYKGKKT CLCGSVKCRG SFT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
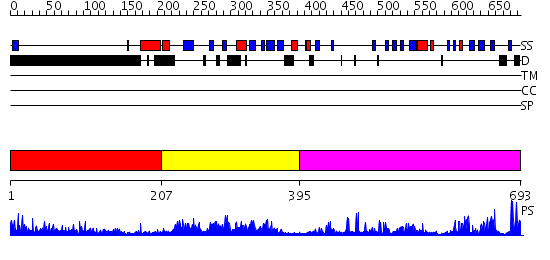
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..206] | 78.223997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [207..394] | 40.958607 | No description for PF02182.8 was found. No confident structure predictions are available. | |
| 3 | View Details | [395..693] | 70.154902 | No description for 2o8jA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)