













| General Information: |
|
| Name(s) found: |
SUVH6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 790 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
histone methylation
[IDA]
|
| Molecular Function: |
methyl-CpNpG binding
[IDA]
methyl-CpNpN binding [IDA] methyltransferase activity [IDA] methyl-CpG binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEMGVMENLM VHTEISKVKS QSNGEVEKRG VSVLENGGVC KLDRMSGLKF KRRKVFAVRD 60
61 FPPGCGSRAM EVKIACENGN VVEDVKVVES LVKEEESLGQ RDASENVSDI RMAEPVEVQP 120
121 LRICLPGGDV VRDLSVTAGD ECSNSEQIVA GSGVSSSSGT ENIVRDIVVY ADESSLGMDN 180
181 LDQTQPLEIE MSDVAVAKPR LVAGRKKAKK GIACHSSLKV VSREFGEGSR KKKSKKNLYW 240
241 RDRESLDSPE QLRILGVGTS SGSSSGDSSR NKVKETLRLF HGVCRKILQE DEAKPEDQRR 300
301 KGKGLRIDFE ASTILKRNGK FLNSGVHILG EVPGVEVGDE FQYRMELNIL GIHKPSQAGI 360
361 DYMKYGKAKV ATSIVASGGY DDHLDNSDVL TYTGQGGNVM QVKKKGEELK EPEDQKLITG 420
421 NLALATSIEK QTPVRVIRGK HKSTHDKSKG GNYVYDGLYL VEKYWQQVGS HGMNVFKFQL 480
481 RRIPGQPELS WVEVKKSKSK YREGLCKLDI SEGKEQSPIS AVNEIDDEKP PLFTYTVKLI 540
541 YPDWCRPVPP KSCCCTTRCT EAEARVCACV EKNGGEIPYN FDGAIVGAKP TIYECGPLCK 600
601 CPSSCYLRVT QHGIKLPLEI FKTKSRGWGV RCLKSIPIGS FICEYVGELL EDSEAERRIG 660
661 NDEYLFDIGN RYDNSLAQGM SELMLGTQAG RSMAEGDESS GFTIDAASKG NVGRFINHSC 720
721 SPNLYAQNVL YDHEDSRIPH VMFFAQDNIP PLQELCYDYN YALDQVRDSK GNIKQKPCFC 780
781 GAAVCRRRLY |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
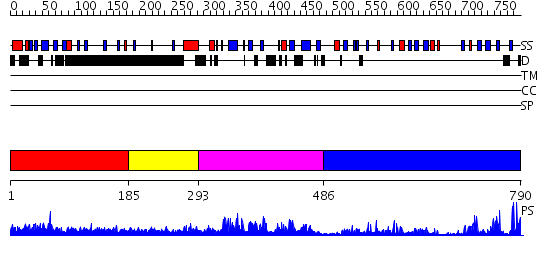
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..184] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [185..292] | 3.69897 | No description for 2e6sA was found. | |
| 3 | View Details | [293..485] | 2.085986 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [486..790] | 49.221849 | No description for 2igqA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)