













| General Information: |
|
| Name(s) found: |
SUVH1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 670 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
regulation of gene expression, epigenetic
[TAS]
|
| Molecular Function: |
histone methyltransferase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERNGGHYTD KTRVLDIKPL RTLRPVFPSG NQAPPFVCAP PFGPFPPGFS SFYPFSSSQA 60
61 NQHTPDLNQA QYPPQHQQPQ NPPPVYQQQP PQHASEPSLV TPLRSFRSPD VSNGNAELEG 120
121 STVKRRIPKK RPISRPENMN FESGINVADR ENGNRELVLS VLMRFDALRR RFAQLEDAKE 180
181 AVSGIIKRPD LKSGSTCMGR GVRTNTKKRP GIVPGVEIGD VFFFRFEMCL VGLHSPSMAG 240
241 IDYLVVKGET EEEPIATSIV SSGYYDNDEG NPDVLIYTGQ GGNADKDKQS SDQKLERGNL 300
301 ALEKSLRRDS AVRVIRGLKE ASHNAKIYIY DGLYEIKESW VEKGKSGHNT FKYKLVRAPG 360
361 QPPAFASWTA IQKWKTGVPS RQGLILPDMT SGVESIPVSL VNEVDTDNGP AYFTYSTTVK 420
421 YSESFKLMQP SFGCDCANLC KPGNLDCHCI RKNGGDFPYT GNGILVSRKP MIYECSPSCP 480
481 CSTCKNKVTQ MGVKVRLEVF KTANRGWGLR SWDAIRAGSF ICIYVGEAKD KSKVQQTMAN 540
541 DDYTFDTTNV YNPFKWNYEP GLADEDACEE MSEESEIPLP LIISAKNVGN VARFMNHSCS 600
601 PNVFWQPVSY ENNSQLFVHV AFFAISHIPP MTELTYDYGV SRPSGTQNGN PLYGKRKCFC 660
661 GSAYCRGSFG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
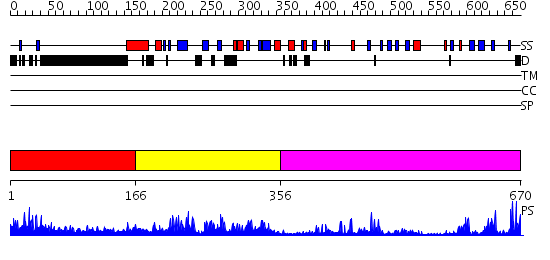
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..165] | 14.045997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [166..355] | 63.721246 | No description for PF02182.8 was found. No confident structure predictions are available. | |
| 3 | View Details | [356..670] | 47.69897 | No description for 2o8jA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)