













| General Information: |
|
| Name(s) found: |
SUT32_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 646 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
integral to membrane [IEA] |
| Biological Process: |
sulfate transport
[IEA]
transport [IEA] |
| Molecular Function: |
sulfate transmembrane transporter activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSKRASQYH QVEIPPPQPF LKSLKNTLNE ILFADDPFRR IRNESKTSKK IELGLRHVFP 60
61 ILEWARGYSL EYLKSDVISG ITIASLAIPQ GISYAQLANL PPILGLYSSL VPPLVYAIMG 120
121 SSRDLAVGTV AVASLLTAAM LGKEVNAVVN PKLYLHLAFT ATFFAGLMQT CLGLLRLGFV 180
181 VEILSHAAIV GFMGGAATVV CLQQLKGLLG LHHFTHSTDI VTVLRSIFSQ SHMWRWESGV 240
241 LGCCFLIFLL TTKYISKKRP KLFWISAMSP LVSVIFGTIF LYFLHDQFHG IQFIGELKKG 300
301 INPPSITHLV FTPPYVMLAL KVGIITGVIA LAEGIAVGRS FAMYKNYNID GNKEMIAFGM 360
361 MNILGSFSSC YLTTGPFSRS AVNYNAGCKT ALSNVVMAVA VAVTLLFLTP LFFYTPLVVL 420
421 SSIIIAAMLG LVDYEAAIHL WKLDKFDFFV CLSAYLGVVF GTIEIGLILS VGISVMRLVL 480
481 FVGRPKIYVM GNIQNSEIYR NIEHYPQAIT RSSLLILHID GPIYFANSTY LRDRIGRWID 540
541 EEEDKLRTSG DISLQYIVLD MSAVGNIDTS GISMLEELNK ILGRRELKLV IANPGAEVMK 600
601 KLSKSTFIES IGKERIYLTV AEAVAACDFM LHTAKPDSPV PEFNNV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
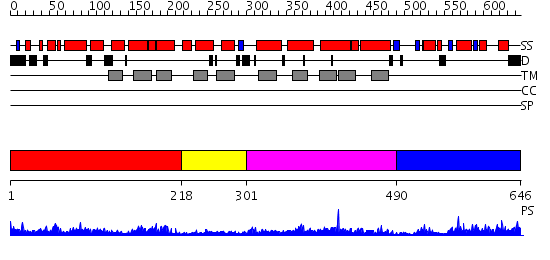
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..217] | 6.894996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [218..300] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [301..489] | 7.908994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [490..646] | 11.0 | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS, NMR, 24 STRUCTURES |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|