













| General Information: |
|
| Name(s) found: |
SPE1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 702 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to oxidative stress
[IEP]
response to cold [IMP] response to salt stress [IMP] seed development [IGI] |
| Molecular Function: |
arginine decarboxylase activity
[NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPALAFVDTP IDTFSSIFTP SSVSTAVVDG SCHWSPSLSS SLYRIDGWGA PYFAANSSGN 60
61 ISVRPHGSNT LPHQDIDLMK VVKKVTDPSG LGLQLPLIVR FPDVLKNRLE CLQSAFDYAI 120
121 QSQGYDSHYQ GVYPVKCNQD RFIIEDIVEF GSGFRFGLEA GSKPEILLAM SCLCKGNPEA 180
181 FLVCNGFKDS EYISLALFGR KLELNTVIVL EQEEELDLVI DLSQKMNVRP VIGLRAKLRT 240
241 KHSGHFGSTS GEKGKFGLTT VQILRVVRKL SQVGMLDCLQ LLHFHIGSQI PSTALLSDGV 300
301 AEAAQLYCEL VRLGAHMKVI DIGGGLGIDY DGSKSGESDL SVAYSLEEYA AAVVASVRFV 360
361 CDQKSVKHPV ICSESGRAIV SHHSVLIFEA VSAGQQHETP TDHQFMLEGY SEEVRGDYEN 420
421 LYGAAMRGDR ESCLLYVDQL KQRCVEGFKE GSLGIEQLAG VDGLCEWVIK AIGASDPVLT 480
481 YHVNLSVFTS IPDFWGIDQL FPIVPIHKLD QRPAARGILS DLTCDSDGKI NKFIGGESSL 540
541 PLHEMDNNGC SGGRYYLGMF LGGAYEEALG GVHNLFGGPS VVRVLQSDGP HGFAVTRAVM 600
601 GQSSADVLRA MQHEPELMFQ TLKHRAEEPR NNNNKACGDK GNDKLVVASC LAKSFNNMPY 660
661 LSMETSTNAL TAAVNNLGVY YCDEAAAGGG GKGKDENWSY FG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
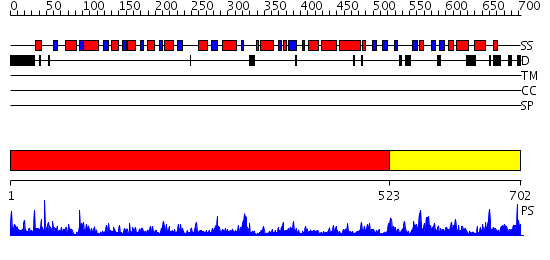
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..522] | 83.221849 | No description for 2p3eA was found. | |
| 2 | View Details | [523..702] | 2.107949 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)