













| General Information: |
|
| Name(s) found: |
MYC2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 623 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
response to abscisic acid stimulus
[IEP]
regulation of transcription from RNA polymerase II promoter in response to oxidative stress [IMP] positive regulation of flavonoid biosynthetic process [IEP] response to wounding [IEP][TAS] response to desiccation [IEP] positive regulation of transcription [IDA] response to chitin [IEP] jasmonic acid mediated signaling pathway [IMP] regulation of transcription factor activity [IMP] |
| Molecular Function: |
DNA binding
[IDA][ISS]
transcription factor activity [ISS] transcription activator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTDYRLQPTM NLWTTDDNAS MMEAFMSSSD ISTLWPPAST TTTTATTETT PTPAMEIPAQ 60
61 AGFNQETLQQ RLQALIEGTH EGWTYAIFWQ PSYDFSGASV LGWGDGYYKG EEDKANPRRR 120
121 SSSPPFSTPA DQEYRKKVLR ELNSLISGGV APSDDAVDEE VTDTEWFFLV SMTQSFACGA 180
181 GLAGKAFATG NAVWVSGSDQ LSGSGCERAK QGGVFGMHTI ACIPSANGVV EVGSTEPIRQ 240
241 SSDLINKVRI LFNFDGGAGD LSGLNWNLDP DQGENDPSMW INDPIGTPGS NEPGNGAPSS 300
301 SSQLFSKSIQ FENGSSSTIT ENPNLDPTPS PVHSQTQNPK FNNTFSRELN FSTSSSTLVK 360
361 PRSGEILNFG DEGKRSSGNP DPSSYSGQTQ FENKRKRSMV LNEDKVLSFG DKTAGESDHS 420
421 DLEASVVKEV AVEKRPKKRG RKPANGREEP LNHVEAERQR REKLNQRFYA LRAVVPNVSK 480
481 MDKASLLGDA IAYINELKSK VVKTESEKLQ IKNQLEEVKL ELAGRKASAS GGDMSSSCSS 540
541 IKPVGMEIEV KIIGWDAMIR VESSKRNHPA ARLMSALMDL ELEVNHASMS VVNDLMIQQA 600
601 TVKMGFRIYT QEQLRASLIS KIG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
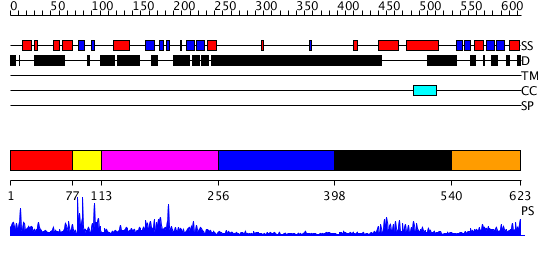
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [77..112] | 2.076989 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [113..255] | 1.13 | Crystal structure of an hypothetical protein | |
| 4 | View Details | [256..397] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [398..539] | 16.39794 | Myc prot-oncogene protein | |
| 6 | View Details | [540..623] | 1.17 | ACT domain protein from Streptococcus pneumoniae |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)