













| General Information: |
|
| Name(s) found: |
PSMD6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 387 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS]
membrane [IDA] proteasome regulatory particle, lid subcomplex [ISS] plasma membrane [IDA] |
| Biological Process: |
protein catabolic process
[TAS]
ubiquitin-dependent protein catabolic process [TAS] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGGAEGSQQ PHLILANKLF LLTHPDVPDI EKVQLKSEVL DFIRSHGMAP LYETLIASSV 60
61 LDLDQSLLES MRAANEEELK KLDEKIADAE ENLGESEVRE AHLAKALYFI RISDKEKALE 120
121 QLKLTEGKTV AVGQKMDVVF YTLQLAFFYM DFDLVSKSID KAKKLFEEGG DWERKNRLKV 180
181 YEGLYCMSTR NFKKAASLFL DSISTFTTYE IFPYETFIFY TVLTSIITLD RVSLKQKVVD 240
241 APEILTVLGK IPFLSEFLNS LYECQYKAFF SAFAGMAVQI KYDRYLYPHF RFYMREVRTV 300
301 VYSQFLESYK SVTVEAMAKA FGVSVDFIDQ ELSRFIAAGK LHCKIDKVAG VLETNRPDAK 360
361 NALYQATIKQ GDFLLNRIQK LSRVIDL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
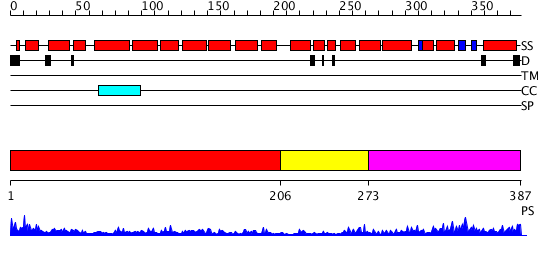
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..205] | 6.09691 | Crystal structure of an 8 repeat consensus TPR superhelix (trigonal crystal form) | |
| 2 | View Details | [206..272] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [273..387] | 10.69897 | Solution structure of the PCI domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)