













| General Information: |
|
| Name(s) found: |
POT4_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 789 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
|
| Biological Process: |
potassium ion transport
[ISS]
response to external stimulus [IEP] |
| Molecular Function: |
potassium ion transmembrane transporter activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPAESGVSP RRNPSQLSWM NLSSNLILAY QSFGVVYGDL STSPLYVFPS TFIGKLHKHH 60
61 NEDAVFGAFS LIFWTLTLIP LLKYLLVLLS ADDNGEGGTF ALYSLLCRHA KLSLLPNQQA 120
121 ADEELSAYKF GPSTDTVTSS PFRTFLEKHK RLRTALLLVV LFGAAMVIGD GVLTPALSVL 180
181 SSLSGLQATE KNVTDGELLV LACVILVGLF ALQHCGTHRV AFMFAPIVII WLISIFFIGL 240
241 YNIIRWNPKI IHAVSPLYII KFFRVTGQDG WISLGGVLLS VTGTEAMFAN LGHFTSVSIR 300
301 VAFAVVVYPC LVVQYMGQAA FLSKNLGSIP NSFYDSVPDP VFWPVFVIAT LAAIVGSQAV 360
361 ITTTFSIIKQ CHALGCFPRI KVVHTSKHIY GQIYIPEINW ILMILTLAMA IGFRDTTLIG 420
421 NAYGIACMVV MFITTFFMAL VIVVVWQKSC FLAALFLGTL WIIEGVYLSA ALMKVTEGGW 480
481 VPFVLTFIFM IAMYVWHYGT RRKYSFDLHN KVSLKWLLGL GPSLGIVRVP GIGLVYSELA 540
541 TGVPAIFSHF VTNLPAFHKV VVFVCVKSVP VPHVSPEERF LIGRVCPKPY RMYRCIVRYG 600
601 YKDIQREDGD FENQLVQSIA EFIQMEASDL QSSASESQSN DGRMAVLSSQ KSLSNSILTV 660
661 SEVEEIDYAD PTIQSSKSMT LQSLRSVYED EYPQGQVRRR HVRFQLTASS GGMGSSVREE 720
721 LMDLIRAKEA GVAYIMGHSY VKSRKSSSWL KKMAIDIGYS FLRKNCRGPA VALNIPHISL 780
781 IEVGMIYYV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
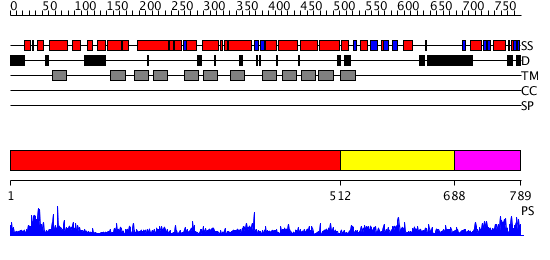
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..511] | 1.01 | Crystal structure of LEUTAA, a bacterial homolog of Na+/Cl--dependent neurotransmitter transporters | |
| 2 | View Details | [512..687] | 1.213995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [688..789] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|