













| General Information: |
|
| Name(s) found: |
PMG2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 560 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apoplast
[IDA]
cytosol [IDA] |
| Biological Process: |
response to cadmium ion
[IEP]
|
| Molecular Function: |
catalytic activity
[IEA]
metal ion binding [IEA] phosphoglycerate mutase activity [IEA] manganese ion binding [IEA] 2,3-bisphosphoglycerate-independent phosphoglycerate mutase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSSGDVNWK LADHPKLPKG KTIGLIVLDG WGESDPDQYN CIHKAPTPAM DSLKDGKPDT 60
61 WRLIKAHGTA VGLPSEDDMG NSEVGHNALG AGRIYAQGAK LVDLALASGK IYEDEGFKYI 120
121 SQSFEKGTVH LIGLLSDGGV HSRLDQVQLL LKGFAERGAK RIRVHILTDG RDVLDGSSVG 180
181 FVETLEADLA ALRAKGVDAQ VASGGGRMYV TMDRYENDWS VVKRGWDAQV LGEAPHKFKS 240
241 ALEAVKTLRA EPGANDQYLP SFVIVDDNGK AVGPIVDGDA VVTFNFRADR MVMHAKALEY 300
301 KDFDKFDRVR VPDIRYAGML QYDGELKLPS RYLVSPPLID RTSGEYLAHN GVRTFACSET 360
361 VKFGHVTFFW NGNRSGYFNE KLEEYVEIPS DSGISFNVQP KMKALEIAEK ARDAILSGKF 420
421 DQVRVNLPNG DMVGHTGDIE ATVVACEAAD RAVRTILDAI EQVGGIYVVT ADHGNAEDMV 480
481 KRDKSGKPAL DKEGNLQILT SHTLKPVPIA IGGPGLSAGV RFRQDIETPG LANVAATVMN 540
541 LHGFVAPSDY ETSLIEVVEK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
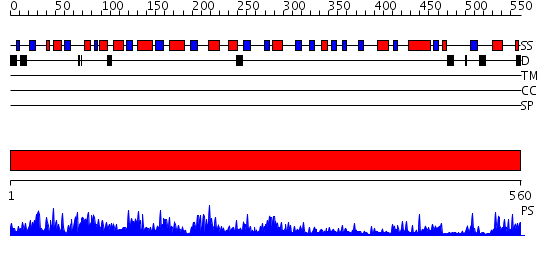
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..560] | 1000.0 | 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain; 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)