













| General Information: |
|
| Name(s) found: |
PGMC2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 585 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytosol [IDA] plasma membrane [IDA] |
| Biological Process: |
carbohydrate metabolic process
[ISS]
response to cadmium ion [IEP] |
| Molecular Function: |
magnesium ion binding
[IEA]
phosphoglucomutase activity [ISS] intramolecular transferase activity, phosphotransferases [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVSFKVSLVS TSPIDGQKPG TSGLRKKVKV FKQPNYLENF VQATFNALTT EKVKGATLVV 60
61 SGDGRYYSEQ AIQIIVKMAA ANGVRRVWVG QNSLLSTPAV SAIIRERVGA DGSKATGAFI 120
121 LTASHNPGGP TEDFGIKYNM ENGGPAPESI TDKIYENTKT IKEYPIAEDL PRVDISTIGI 180
181 TSFEGPEGKF DVEVFDSADD YVKLMKSIFD FESIKKLLSY PKFTFCYDAL HGVAGAYAHR 240
241 IFVEELGAPE SSLLNCVPKE DFGGGHPDPN LTYAKELVAR MGLSKTDDAG GEPPEFGAAA 300
301 DGDADRNMIL GKRFFVTPSD SVAIIAANAV GAIPYFSSGL KGVARSMPTS AALDVVAKNL 360
361 GLKFFEVPTG WKFFGNLMDA GMCSVCGEES FGTGSDHIRE KDGIWAVLAW LSILAHKNKE 420
421 TLDGNAKLVT VEDIVRQHWA TYGRHYYTRY DYENVDATAA KELMGLLVKL QSSLPEVNKI 480
481 IKGIHPEVAN VASADEFEYK DPVDGSVSKH QGIRYLFEDG SRLVFRLSGT GSEGATIRLY 540
541 IEQYEKDASK IGRDSQDALG PLVDVALKLS KMQEFTGRSS PTVIT |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
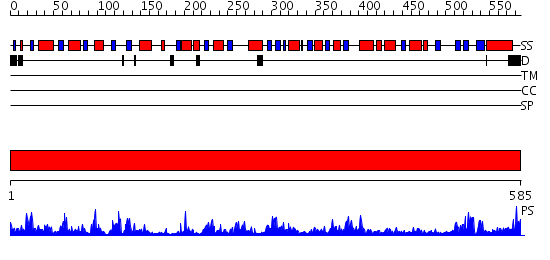
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..585] | 152.0 | Phosphoglucomutase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)