













| General Information: |
|
| Name(s) found: |
PLDZ1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1096 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
root development
[IGI]
[NOT]cellular response to phosphate starvation [IEP] |
| Molecular Function: |
phospholipase D activity
[ISS][TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASEQLMSPA SGGGRYFQMQ PEQFPSMVSS LFSFAPAPTQ ETNRIFEELP KAVIVSVSRP 60
61 DAGDISPVLL SYTIECQYKQ FKWQLVKKAS QVFYLHFALK KRAFIEEIHE KQEQVKEWLQ 120
121 NLGIGDHPPV VQDEDADEVP LHQDESAKNR DVPSSAALPV IRPLGRQQSI SVRGKHAMQE 180
181 YLNHFLGNLD IVNSREVCRF LEVSMLSFSP EYGPKLKEDY IMVKHLPKFS KSDDDSNRCC 240
241 GCCWFCCCND NWQKVWGVLK PGFLALLEDP FDAKLLDIIV FDVLPVSNGN DGVDISLAVE 300
301 LKDHNPLRHA FKVTSGNRSI RIRAKNSAKV KDWVASINDA ALRPPEGWCH PHRFGSYAPP 360
361 RGLTDDGSQA QWFVDGGAAF AAIAAAIENA KSEIFICGWW VCPELYLRRP FDPHTSSRLD 420
421 NLLENKAKQG VQIYILIYKE VALALKINSV YSKRRLLGIH ENVRVLRYPD HFSSGVYLWS 480
481 HHEKLVIVDN QVCFIGGLDL CFGRYDTFEH KVGDNPSVTW PGKDYYNPRE SEPNTWEDAL 540
541 KDELERKKHP RMPWHDVHCA LWGPPCRDVA RHFVQRWNYA KRNKAPYEDS IPLLMPQHHM 600
601 VIPHYMGRQE ESDIESKKEE DSIRGIRRDD SFSSRSSLQD IPLLLPHEPV DQDGSSGGHK 660
661 ENGTNNRNGP FSFRKSKIEP VDGDTPMRGF VDDRNGLDLP VAKRGSNAID SEWWETQDHD 720
721 YQVGSPDETG QVGPRTSCRC QIIRSVSQWS AGTSQVEESI HSAYRSLIDK AEHFIYIENQ 780
781 FFISGLSGDD TVKNRVLEAL YKRILRAHNE KKIFRVVVVI PLLPGFQGGI DDSGAASVRA 840
841 IMHWQYRTIY RGHNSILTNL YNTIGVKAHD YISFYGLRAY GKLSEDGPVA TSQVYVHSKI 900
901 MIVDDRAALI GSANINDRSL LGSRDSEIGV LIEDTELVDS RMAGKPWKAG KFSSSLRLSL 960
961 WSEHLGLRTG EIDQIIDPVS DSTYKEIWMA TAKTNTMIYQ DVFSCVPNDL IHSRMAFRQS1020
1021 LSYWKEKLGH TTIDLGIAPE KLESYHNGDI KRSDPMDRLK AIKGHLVSFP LDFMCKEDLR1080
1081 PVFNESEYYA SPQVFH |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
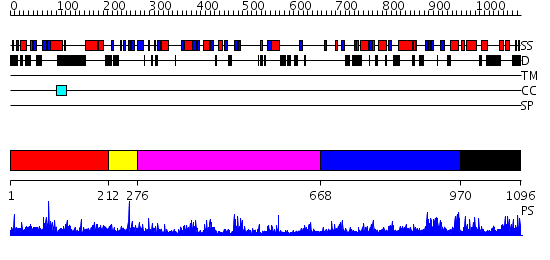
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..211] | 2.27 | Crystal structure of the yeast PX-doamin protein Grd19p (sorting nexin3) complexed to phosphatidylinosytol-3-phosphate. | |
| 2 | View Details | [212..275] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [276..667] | 22.522879 | Phospholipase D | |
| 4 | View Details | [668..969] | 4.41 | Tungstate-inhibited phospholipase D from Streptomyces sp. strain PMF. | |
| 5 | View Details | [970..1096] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 |
|
||||||
| 4 |
|
||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)