













| General Information: |
|
| Name(s) found: |
PSD3B_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 487 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IDA]
proteasome regulatory particle, lid subcomplex [ISS] plasma membrane [IDA] |
| Biological Process: |
protein catabolic process
[TAS]
ubiquitin-dependent protein catabolic process [TAS] |
| Molecular Function: |
enzyme regulator activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTQDVEMKDN QTPTQSVVSA PTSTLQNLKE IAALIDTGSY TKEVRRIARA VRLTVGLRRK 60
61 LTGSVISSFL DFALVPGSEA HTRLSSFVPK SDEHDMEVDT ASSNSQAPPK HLPAELEIYC 120
121 YFIVLLFLID QKKYNEAKVC STASIARLKS VNRRTVDVIA SKLYFYYSLS YELTNDLAEI 180
181 RSTLLALHHS ATLRHDELGQ ETLLNLLLRN YLHYNLYDQA EKLRSKAPRF EAHSNQQFCR 240
241 YLFYLGKIRT IQLEYTDAKE SLLQAARKAP VASLGFRIQC NKWAIIVRLL LGEIPERSIF 300
301 TQKGMEKTLR PYFELTNAVR IGDLELFGKI QEKFAKTFAE DRTHNLIVRL RHNVIRTGLR 360
361 NISISYSRIS LQDVAQKLRL NSANPVADAE SIVAKAIRDG AIDATIDHKN GCMVSKETGD 420
421 IYSTNEPQTA FNSRIAFCLN MHNEAVRALR FPPNTHREKE SEEKRREMKQ QEEELAKYMA 480
481 EEDDDDF |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
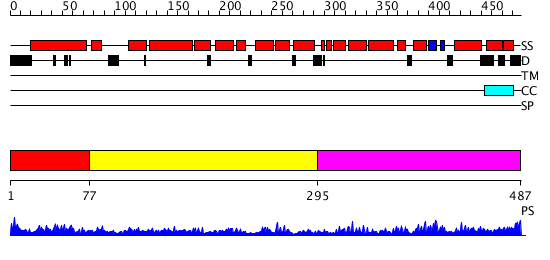
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | 1.141994 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [77..294] | 5.045757 | Crystal structure of an 8 repeat consensus TPR superhelix (trigonal crystal form) | |
| 3 | View Details | [295..487] | 8.69897 | Solution structure of the PCI domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)