













| General Information: |
|
| Name(s) found: |
P5CS2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 726 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
cytoplasm [IDA] |
| Biological Process: |
hyperosmotic salinity response
[IEP]
response to abscisic acid stimulus [IEP] embryonic development ending in seed dormancy [IMP] proline biosynthetic process [IMP][TAS] |
| Molecular Function: |
glutamate 5-kinase activity
[IEA]
catalytic activity [IEA] oxidoreductase activity [IEA] glutamate-5-semialdehyde dehydrogenase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEIDRSRAF AKDVKRIVVK VGTAVVTGKG GRLALGRLGA ICEQLAELNS DGFEVILVSS 60
61 GAVGLGRQRL RYRQLVNSSF ADLQKPQMEL DGKACAGVGQ SSLMAYYETM FDQLDVTVAQ 120
121 MLVTDSSFRD KDFRKQLSET VKAMLRMRVI PVFNENDAIS TRRAPYKDST GIFWDNDSLA 180
181 ALLSLELKAD LLILLSDVEG LYTGPPSDST SKLIHTFIKE KHQDEITFGE KSKLGRGGMT 240
241 AKVKAAVNAA YGGVPVIITS GYAAENISKV LRGLRVGTLF HQDAHLWAPV VDTTSRDMAV 300
301 AARESSRKLQ ALSSEDRKQI LHDIANALEV NEKTIKAEND LDVAAAQEAG YEESLVARLV 360
361 MKPGKISSLA ASVRQLAEME DPIGRVLKKT QVADDLILEK TSSPIGVLLI VFESRPDALV 420
421 QIASLAIRSG NGLLLKGGKE ARRSNAILHK VITDAIPETV GGKLIGLVTS REEIPDLLKL 480
481 DDVIDLVIPR GSNKLVSQIK NSTKIPVLGH ADGICHVYVD KSGKLDMAKR IVSDAKLDYP 540
541 AACNAMETLL VHKDLEQNGF LDDLIYVLQT KGVTLYGGPR ASAKLNIPET KSFHHEYSSK 600
601 ACTVEIVEDV YGAIDHIHQH GSAHTDCIVT EDSEVAEIFL RQVDSAAVFH NASTRFSDGF 660
661 RFGLGAEVGI STSRIHARGP VGVEGLLTTR WIMRGKGQVV DGDNGIVYTH KDLPVLQRTE 720
721 AVENGI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
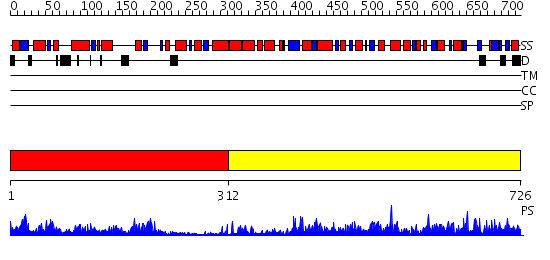
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..311] | 50.39794 | No description for 2j5tA was found. | |
| 2 | View Details | [312..726] | 90.39794 | No description for 2h5gA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)