













| General Information: |
|
| Name(s) found: |
P5CS1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 717 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
cytoplasm [IDA] membrane [IDA] |
| Biological Process: |
root development
[IMP]
response to oxidative stress [IMP] hyperosmotic salinity response [IEP][IMP] response to abscisic acid stimulus [IEP] response to salt stress [IEP] response to desiccation [IEP] proline biosynthetic process [IMP][TAS] response to water deprivation [IEP] |
| Molecular Function: |
delta1-pyrroline-5-carboxylate synthetase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEELDRSRAF ARDVKRIVVK VGTAVVTGKG GRLALGRLGA LCEQLAELNS DGFEVILVSS 60
61 GAVGLGRQRL RYRQLVNSSF ADLQKPQTEL DGKACAGVGQ SSLMAYYETM FDQLDVTAAQ 120
121 LLVNDSSFRD KDFRKQLNET VKSMLDLRVI PIFNENDAIS TRRAPYQDSS GIFWDNDSLA 180
181 ALLALELKAD LLILLSDVEG LYTGPPSDPN SKLIHTFVKE KHQDEITFGD KSRLGRGGMT 240
241 AKVKAAVNAA YAGIPVIITS GYSAENIDKV LRGLRVGTLF HQDARLWAPI TDSNARDMAV 300
301 AARESSRKLQ ALSSEDRKKI LLDIADALEA NVTTIKAENE LDVASAQEAG LEESMVARLV 360
361 MTPGKISSLA ASVRKLADME DPIGRVLKKT EVADGLVLEK TSSPLGVLLI VFESRPDALV 420
421 QIASLAIRSG NGLLLKGGKE ARRSNAILHK VITDAIPETV GGKLIGLVTS REEIPDLLKL 480
481 DDVIDLVIPR GSNKLVTQIK NTTKIPVLGH ADGICHVYVD KACDTDMAKR IVSDAKLDYP 540
541 AACNAMETLL VHKDLEQNAV LNELIFALQS NGVTLYGGPR ASKILNIPEA RSFNHEYCAK 600
601 ACTVEVVEDV YGAIDHIHRH GSAHTDCIVT EDHEVAELFL RQVDSAAVFH NASTRFSDGF 660
661 RFGLGAEVGV STGRIHARGP VGVEGLLTTR WIMRGKGQVV DGDNGIVYTH QDIPIQA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
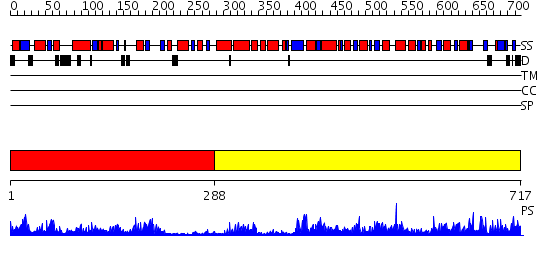
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..287] | 50.522879 | No description for 2j5tA was found. | |
| 2 | View Details | [288..717] | 89.09691 | No description for 2h5gA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)