













| General Information: |
|
| Name(s) found: |
MSH2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 937 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
mismatch repair
[IMP][ISS]
negative regulation of reciprocal meiotic recombination [IMP] |
| Molecular Function: |
ATP binding
[ISS]
protein binding [IDA] mismatched DNA binding [IDA] damaged DNA binding [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEGNFEEQNK LPELKLDAKQ AQGFLSFYKT LPNDTRAVRF FDRKDYYTAH GENSVFIAKT 60
61 YYHTTTALRQ LGSGSNALSS VSISRNMFET IARDLLLERN DHTVELYEGS GSNWRLVKTG 120
121 SPGNIGSFED VLFANNEMQD TPVVVSIFPS FHDGRCVIGM AYVDLTRRVL GLAEFLDDSR 180
181 FTNLESSLIA LGAKECIFPA ESGKSNECKS LYDSLERCAV MITERKKHEF KGRDLDSDLK 240
241 RLVKGNIEPV RDLVSGFDLA TPALGALLSF SELLSNEDNY GNFTIRRYDI GGFMRLDSAA 300
301 MRALNVMESK TDANKNFSLF GLMNRTCTAG MGKRLLHMWL KQPLVDLNEI KTRLDIVQCF 360
361 VEEAGLRQDL RQHLKRISDV ERLLRSLERR RGGLQHIIKL YQSTIRLPFI KTAMQQYTGE 420
421 FASLISERYL KKLEALSDQD HLGKFIDLVE CSVDLDQLEN GEYMISSSYD TKLASLKDQK 480
481 ELLEQQIHEL HKKTAIELDL QVDKALKLDK AAQFGHVFRI TKKEEPKIRK KLTTQFIVLE 540
541 TRKDGVKFTN TKLKKLGDQY QSVVDDYRSC QKELVDRVVE TVTSFSEVFE DLAGLLSEMD 600
601 VLLSFADLAA SCPTPYCRPE ITSSDAGDIV LEGSRHPCVE AQDWVNFIPN DCRLMRGKSW 660
661 FQIVTGPNMG GKSTFIRQVG VIVLMAQVGS FVPCDKASIS IRDCIFARVG AGDCQLRGVS 720
721 TFMQEMLETA SILKGASDKS LIIIDELGRG TSTYDGFGLA WAICEHLVQV KRAPTLFATH 780
781 FHELTALAQA NSEVSGNTVG VANFHVSAHI DTESRKLTML YKVEPGACDQ SFGIHVAEFA 840
841 NFPESVVALA REKAAELEDF SPSSMIINNE ESGKRKSRED DPDEVSRGAE RAHKFLKEFA 900
901 AIPLDKMELK DSLQRVREMK DELEKDAADC HWLRQFL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
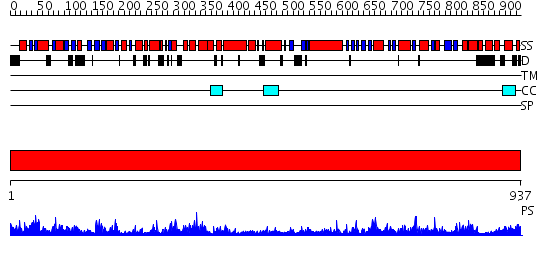
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..937] | 1000.0 | No description for 2o8bA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)